Fitting MF (or other) datasets
12/09/22
For the basic (default) case, using AF data, see the basic demo notebook and PEMtk fitting setup & batch run demo notebook. This notebook is based on the latter, but demonstrates additionally:
Switching the fit dataset to use MF data as a function of polarization (as opposed to AF data as a function of alignment/time in the earlier demo case).
Switching the backend to calculate MFPADs (see the backends notebook for more).
Running fits.
Note that the methods here should be general, allowing for fitting of MF or AF/LF data vs. alignment, time, energy or polarization geometry depending on the dataset.
Setup
Here we’ll use the demo script, as per the PEMtk fitting setup & batch run demo notebook, then replace some of the defaults later. Alternatively, the MF case could be setup from scratch following the basic demo notebook.
[1]:
# Import & set paths
import pemtk
from pemtk.fit.fitClass import pemtkFit
from pathlib import Path
# Path for demo script
demoPath = Path(pemtk.__file__).parent.parent/Path('demos','fitting')
# Run demo script to configure workspace
%run {demoPath/"setup_fit_demo.py"}
*** ePSproc installation not found, setting for local copy.
* Setting plotter defaults with epsproc.basicPlotters.setPlotters(). Run directly to modify, or change options in local env.
* Set Holoviews with bokeh.
C:\Users\femtolab\.conda\envs\ePSdev\lib\site-packages\xyzpy\plot\xyz_cmaps.py:6: MatplotlibDeprecationWarning:
The revcmap function was deprecated in Matplotlib 3.2 and will be removed two minor releases later. Use Colormap.reversed() instead.
return LinearSegmentedColormap(name, cm.revcmap(cmap._segmentdata))
*** Setting up demo fitting workspace and main `data` class object...
For more details see https://pemtk.readthedocs.io/en/latest/fitting/PEMtk_fitting_basic_demo_030621-full.html
To use local source code, pass the parent path to this script at run time, e.g. "setup_fit_demo ~/github"
* Loading packages...
* Set Holoviews with bokeh.
* Loading demo matrix element data from D:\code\github\ePSproc\data\photoionization\n2_multiorb...
*** Job subset details
Key: subset
No 'job' info set for self.data[subset].
*** Job orb6 details
Key: orb6
Dir D:\code\github\ePSproc\data\photoionization\n2_multiorb, 1 file(s).
{ 'batch': 'ePS n2, batch n2_1pu_0.1-50.1eV, orbital A2',
'event': ' N2 A-state (1piu-1)',
'orbE': -17.096913836366,
'orbLabel': '1piu-1'}
*** Job orb5 details
Key: orb5
Dir D:\code\github\ePSproc\data\photoionization\n2_multiorb, 1 file(s).
{ 'batch': 'ePS n2, batch n2_3sg_0.1-50.1eV, orbital A2',
'event': ' N2 X-state (3sg-1)',
'orbE': -17.341816310545997,
'orbLabel': '3sg-1'}
* Loading demo ADM data from D:\code\github\ePSproc\data\alignment\N2_ADM_VM_290816.mat...
* Subselecting data...
Subselected from dataset 'orb5', dataType 'matE': 36 from 11016 points (0.33%)
Subselected from dataset 'pol', dataType 'pol': 1 from 3 points (33.33%)
Subselected from dataset 'ADM', dataType 'ADM': 52 from 14764 points (0.35%)
* Calculating AF-BLMs...
Subselected from dataset 'sim', dataType 'AFBLM': 195 from 195 points (100.00%)
*Setting up fit parameters (with constraints)...
Set 6 complex matrix elements to 12 fitting params, see self.params for details.
C:\Users\femtolab\.conda\envs\ePSdev\lib\site-packages\pandas\core\generic.py:3939: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
self._update_inplace(obj)
Auto-setting parameters.
| name | value | initial value | min | max | vary | expression |
|---|---|---|---|---|---|---|
| m_PU_SG_PU_1_n1_1_1 | 1.78461575 | 1.784615753610107 | 1.0000e-04 | 5.00000000 | True | |
| m_PU_SG_PU_1_1_n1_1 | 1.78461575 | 1.784615753610107 | 1.0000e-04 | 5.00000000 | False | m_PU_SG_PU_1_n1_1_1 |
| m_PU_SG_PU_3_n1_1_1 | 0.80290495 | 0.802904951323892 | 1.0000e-04 | 5.00000000 | True | |
| m_PU_SG_PU_3_1_n1_1 | 0.80290495 | 0.802904951323892 | 1.0000e-04 | 5.00000000 | False | m_PU_SG_PU_3_n1_1_1 |
| m_SU_SG_SU_1_0_0_1 | 2.68606212 | 2.686062120382649 | 1.0000e-04 | 5.00000000 | True | |
| m_SU_SG_SU_3_0_0_1 | 1.10915311 | 1.109153108617096 | 1.0000e-04 | 5.00000000 | True | |
| p_PU_SG_PU_1_n1_1_1 | -0.86104140 | -0.8610414024232179 | -3.14159265 | 3.14159265 | False | |
| p_PU_SG_PU_1_1_n1_1 | -0.86104140 | -0.8610414024232179 | -3.14159265 | 3.14159265 | False | p_PU_SG_PU_1_n1_1_1 |
| p_PU_SG_PU_3_n1_1_1 | -3.12044446 | -3.1204444620772467 | -3.14159265 | 3.14159265 | True | |
| p_PU_SG_PU_3_1_n1_1 | -3.12044446 | -3.1204444620772467 | -3.14159265 | 3.14159265 | False | p_PU_SG_PU_3_n1_1_1 |
| p_SU_SG_SU_1_0_0_1 | 2.61122920 | 2.611229196458127 | -3.14159265 | 3.14159265 | True | |
| p_SU_SG_SU_3_0_0_1 | -0.07867828 | -0.07867827542158025 | -3.14159265 | 3.14159265 | True |
*** Setup demo fitting workspace OK.
Default AF case
As shown above, the demo script set some AF fitting defaults. These are also shown in some of the optional settings…
[2]:
data.selOpts
[2]:
{'matE': {'thres': 0.01, 'inds': {'Type': 'L', 'Eke': 1.1}},
'slices': {'t': [4, 5, 4]},
'pol': {'inds': {'Labels': 'z'}},
'ADM': {},
'AFBLM': {}}
[3]:
# data.fitOpts['backend'].__name__
data.backend
[3]:
'afblmXprod'
[4]:
# Data is set as AFBLMs, vs. time
data.data['subset']['AFBLM']
[4]:
- Labels: 1
- t: 13
- BLM: 15
- 0j (1.668823121150882+5.788281730622143e-18j) ... 0j
array([[[ 0.00000000e+00+0.00000000e+00j, 1.66882312e+00+5.78828173e-18j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 9.25154735e-01-2.04255472e-18j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, -1.45626494e-01-2.50162312e-18j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 9.05679279e-03+2.69005757e-19j, 0.00000000e+00+0.00000000e+00j], [ 0.00000000e+00+0.00000000e+00j, 1.65939046e+00-7.23129056e-18j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 9.28209270e-01-1.20305969e-17j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, -1.35884180e-01+7.60995910e-18j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 7.87360160e-03-5.26232236e-19j, 0.00000000e+00+0.00000000e+00j], [ 0.00000000e+00+0.00000000e+00j, 1.60819595e+00+5.37946903e-19j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 9.44773203e-01+2.80576809e-17j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, -8.24684513e-02-1.29331051e-18j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 1.24796733e-03+1.99111244e-19j, 0.00000000e+00+0.00000000e+00j], [ 0.00000000e+00+0.00000000e+00j, 1.52326324e+00-1.50526506e-18j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 9.72371636e-01+7.27400639e-19j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 4.93224211e-04+1.20678198e-18j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, -5.69662854e-03-1.12782141e-19j, 0.00000000e+00+0.00000000e+00j], [ 0.00000000e+00+0.00000000e+00j, 1.43634451e+00+9.52773406e-18j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 1.00104159e+00+2.33569626e-17j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 6.53535497e-02-2.06092931e-18j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 1.13815711e-03-3.50093184e-18j, 0.00000000e+00+0.00000000e+00j], [ 0.00000000e+00+0.00000000e+00j, 1.38620024e+00+9.20243198e-19j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 1.01780024e+00-1.33980425e-18j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 9.28414140e-02+2.05936950e-18j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 1.15029998e-02-1.86868078e-18j, 0.00000000e+00+0.00000000e+00j], [ 0.00000000e+00+0.00000000e+00j, 1.39472246e+00-4.45373740e-18j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 1.01490490e+00+1.74312370e-18j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 9.03441438e-02+2.26699795e-18j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 8.28715378e-03+9.94032689e-19j, 0.00000000e+00+0.00000000e+00j], [ 0.00000000e+00+0.00000000e+00j, 1.45369338e+00+7.91745171e-19j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 9.95369088e-01+2.73033702e-17j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 5.02659008e-02+1.13177636e-18j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 9.75429570e-04-6.51669777e-19j, 0.00000000e+00+0.00000000e+00j], [ 0.00000000e+00+0.00000000e+00j, 1.53164115e+00+1.13429074e-17j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 9.69964449e-01-3.96161113e-18j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, -2.24085809e-02-7.42900134e-18j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 5.08531171e-03-3.02631277e-18j, 0.00000000e+00+0.00000000e+00j], [ 0.00000000e+00+0.00000000e+00j, 1.59428225e+00-1.99642906e-18j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 9.49718737e-01+1.56864870e-17j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, -8.90643638e-02-5.46842630e-18j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 1.44722472e-02+1.25126193e-18j, 0.00000000e+00+0.00000000e+00j], [ 0.00000000e+00+0.00000000e+00j, 1.62292950e+00-6.46048690e-18j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 9.40438270e-01+1.76164331e-17j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, -1.18540934e-01-1.18827450e-18j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 1.80790278e-02+3.96263817e-18j, 0.00000000e+00+0.00000000e+00j], [ 0.00000000e+00+0.00000000e+00j, 1.61964948e+00-9.65063099e-18j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 9.41433561e-01-1.12629747e-17j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, -1.11981456e-01+8.46145725e-18j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 1.54232880e-02+1.10625182e-18j, 0.00000000e+00+0.00000000e+00j], [ 0.00000000e+00+0.00000000e+00j, 1.59961793e+00+3.78243863e-18j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 9.47859045e-01-1.46603781e-18j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, -8.83338758e-02-1.35167290e-18j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 1.07442112e-02+1.22430637e-18j, 0.00000000e+00+0.00000000e+00j]]]) - Euler(Labels)object(0.0, 0.0, 0.0)
array([(0.0, 0.0, 0.0)], dtype=object)
- Labels(Labels)<U1'A'
array(['A'], dtype='<U1')
- t(t)float644.018 4.096 4.175 ... 4.884 4.962
- units :
- ps
array([4.017657, 4.096371, 4.175086, 4.253801, 4.332516, 4.41123 , 4.489945, 4.56866 , 4.647375, 4.726089, 4.804804, 4.883519, 4.962233]) - Ehv()float6418.4
array(18.4)
- Type()<U1'L'
array('L', dtype='<U1') - Eke()float641.1
array(1.1)
- SF()complex128(2.2237521+3.6277801j)
array(2.2237521+3.6277801j)
- XSraw(Labels, t)complex128(1.668823121150882+5.788281730622143e-18j) ... (1.5996179261474746+3.782438628664796e-18j)
array([[1.66882312+5.78828173e-18j, 1.65939046-7.23129056e-18j, 1.60819595+5.37946903e-19j, 1.52326324-1.50526506e-18j, 1.43634451+9.52773406e-18j, 1.38620024+9.20243198e-19j, 1.39472246-4.45373740e-18j, 1.45369338+7.91745171e-19j, 1.53164115+1.13429074e-17j, 1.59428225-1.99642906e-18j, 1.6229295 -6.46048690e-18j, 1.61964948-9.65063099e-18j, 1.59961793+3.78243863e-18j]]) - XSrescaled(Labels, t)complex128(5.915823935128086+2.0518924487134524e-17j) ... (5.670497906355172+1.3408395826381391e-17j)
array([[5.91582394+2.05189245e-17j, 5.88238602-2.56342576e-17j, 5.70090622+1.90697212e-18j, 5.39982759-5.33602569e-18j, 5.09170872+3.37749379e-17j, 4.91395189+3.26217720e-18j, 4.9441624 -1.57880880e-17j, 5.15320886+2.80666355e-18j, 5.4295265 +4.02095600e-17j, 5.65158343-7.07715676e-18j, 5.75313528-2.29018298e-17j, 5.74150791-3.42105961e-17j, 5.67049791+1.34083958e-17j]]) - XSiso()complex128(5.36805661023236+0j)
array(5.36805661+0.j)
- BLM(BLM)MultiIndex(l, m)
array([(0, -1), (0, 0), (0, 1), (2, -1), (2, 0), (2, 1), (3, -1), (3, 0), (3, 1), (4, -1), (4, 0), (4, 1), (6, -1), (6, 0), (6, 1)], dtype=object) - l(BLM)int640 0 0 2 2 2 3 3 3 4 4 4 6 6 6
array([0, 0, 0, 2, 2, 2, 3, 3, 3, 4, 4, 4, 6, 6, 6], dtype=int64)
- m(BLM)int64-1 0 1 -1 0 1 -1 0 1 -1 0 1 -1 0 1
array([-1, 0, 1, -1, 0, 1, -1, 0, 1, -1, 0, 1, -1, 0, 1], dtype=int64)
- E :
- 0.1
- Ehv :
- 15.68
- SF :
- (2.1560627+3.741674j)
- Lmax :
- 11
- Cont :
- SU
- Targ :
- SG
- Total :
- SU
- QNs :
- None
- dataType :
- BLM
- file :
- n2_3sg_0.1-50.1eV_A2.inp.out
- fileBase :
- D:\code\github\ePSproc\data\photoionization\n2_multiorb
- fileList :
- n2_3sg_0.1-50.1eV_A2.inp.out
- jobLabel :
- 3sg-1
- EPRX :
- None
- p :
- [0]
- BLMtable :
- None
- BLMtableResort :
- None
- lambdaTerm :
- None
- polProd :
- None
- AFterm :
- None
- thres :
- None
- thresDims :
- Eke
- selDims :
- {}
- sumDims :
- ['mu', 'mup', 'l', 'lp', 'm', 'mp', 'S-Rp']
- sumDimsPol :
- ['P', 'R', 'Rp', 'p']
- symSum :
- True
- degenDrop :
- True
- SFflag :
- False
- SFflagRenorm :
- False
- BLMRenorm :
- 0
- squeeze :
- False
- phaseConvention :
- E
- basisReturn :
- ProductBasis
- verbose :
- 0
- kwargs :
- {'RX': <xarray.DataArray ()> array(quaternion(1, -0, 0, 0), dtype=quaternion) Coordinates: Euler object (0.0, 0.0, 0.0) Labels <U18 'z' Attributes: dataType: Euler}
- matEleSelector :
- <function matEleSelector at 0x00000191D7E657B8>
- degenDict :
- {'it': <xarray.DataArray 'it' (it: 1)> array([1], dtype=int64) Coordinates: Ehv float64 18.4 * it (it) int64 1 Type <U1 'L' Eke float64 1.1 SF complex128 (2.2237521+3.6277801j), 'degenN': array(1, dtype=int64), 'degenFlag': False, 'degenDrop': True, 'selDims': {}}
Set the (MF) data to fit
Here we’ll calculate some MF-\(\beta_{LM}\) parameters to use as the test data. Note this currently needs to be set as self.data['subset']['AFBLM'] for fitting (even if it’s a different datatype).
TODO: add dict key option here for different backends.
[5]:
# For MF case remove the existing AFBLM and update pols (these are set in demo script.)
# And for the polarisation geometries...
data.selOpts['pol'] = {}
data.setSubset(dataKey = 'pol', dataType = 'pol')
# Delete ADMs, since we don't
del(data.data['subset']['ADM'])
Subselected from dataset 'pol', dataType 'pol': 3 from 3 points (100.00%)
[6]:
# Set the backend function, see https://pemtk.readthedocs.io/en/latest/fitting/PEMtk_fitting_backends_demo_010922.html
# 21/08/22 - Change backend in dict
# data.fitOpts = {'backend':data.backends(backend='mfblmXprod')}
# 01/09/22 - Change to direct setting via self.backends
data.backends(backend='mfblmXprod')
[7]:
# Generate some data & update basis etc - now set by main fitting func.
# BLMtest, data.basis = data.afblmMatEfit(backend = ep.mfblmXprod, resetBasis = True) # With backend by object
BLMtest, data.basis = data.afblmMatEfit(resetBasis = True) # 21/08/22 With backend as set in data.fitOpts
data.setData('sim', BLMtest) # Set simulated data to master structure as "sim"
data.setSubset('sim','AFBLM') # Set to 'subset' to use for fitting.
Subselected from dataset 'sim', dataType 'AFBLM': 75 from 75 points (100.00%)
[8]:
# Show the data
data.data['subset']['AFBLM']
[8]:
- Labels: 3
- BLM: 25
- 0j 0j (1+0j) ... 0j (-0.07248766838683887-8.877179106759699e-18j)
array([[ 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 1.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 2.93448657e-01+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 3.60745890e-34+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, -4.74637872e-01+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 1.22430606e-01+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j], [ 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 1.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 5.86599819e-01+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, -6.37672502e-01+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 5.86599819e-01+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, -1.52557403e-01+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 2.69489335e-01+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, -1.52557403e-01+0.00000000e+00j, 7.24876684e-02+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, -1.06111081e-01+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 7.24876684e-02+0.00000000e+00j], [ 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 1.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, -5.86599819e-01+7.81830424e-17j, 0.00000000e+00+0.00000000e+00j, -6.37672502e-01+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, -5.86599819e-01-7.81830424e-17j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 1.52557403e-01+5.92656783e-18j, 0.00000000e+00+0.00000000e+00j, 2.69489335e-01+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, 1.52557403e-01-5.92656783e-18j, -7.24876684e-02+8.87717911e-18j, 0.00000000e+00+0.00000000e+00j, -1.06111081e-01+0.00000000e+00j, 0.00000000e+00+0.00000000e+00j, -7.24876684e-02-8.87717911e-18j]]) - Euler(Labels)object(0.0, 0.0, 0.0) ... (1.5707963267948966, 1.5707963267948966, 0.0)
array([(0.0, 0.0, 0.0), (0.0, 1.5707963267948966, 0.0), (1.5707963267948966, 1.5707963267948966, 0.0)], dtype=object) - Labels(Labels)<U18'z' 'x' 'y'
array(['z', 'x', 'y'], dtype='<U18')
- Ehv()float6418.4
array(18.4)
- Type()<U1'L'
array('L', dtype='<U1') - Eke()float641.1
array(1.1)
- SF()complex128(2.2237521+3.6277801j)
array(2.2237521+3.6277801j)
- XS(Labels)complex128(2.3823329246612026+0j) ... (1.0802847552102302+0j)
array([2.38233292+0.j, 1.08028476+0.j, 1.08028476+0.j])
- BLM(BLM)MultiIndex(l, m)
array([(0, -2), (0, -1), (0, 0), (0, 1), (0, 2), (2, -2), (2, -1), (2, 0), (2, 1), (2, 2), (3, -2), (3, -1), (3, 0), (3, 1), (3, 2), (4, -2), (4, -1), (4, 0), (4, 1), (4, 2), (6, -2), (6, -1), (6, 0), (6, 1), (6, 2)], dtype=object) - l(BLM)int640 0 0 0 0 2 2 2 ... 4 4 4 6 6 6 6 6
array([0, 0, 0, 0, 0, 2, 2, 2, 2, 2, 3, 3, 3, 3, 3, 4, 4, 4, 4, 4, 6, 6, 6, 6, 6], dtype=int64) - m(BLM)int64-2 -1 0 1 2 -2 -1 ... 2 -2 -1 0 1 2
array([-2, -1, 0, 1, 2, -2, -1, 0, 1, 2, -2, -1, 0, 1, 2, -2, -1, 0, 1, 2, -2, -1, 0, 1, 2], dtype=int64)
- E :
- 0.1
- Ehv :
- 15.68
- SF :
- (2.1560627+3.741674j)
- Lmax :
- 11
- Cont :
- SU
- Targ :
- SG
- Total :
- SU
- QNs :
- ['m', 'l', 'mu', 'ip', 'it', 'Value']
- dataType :
- BLM
- file :
- n2_3sg_0.1-50.1eV_A2.inp.out
- fileBase :
- D:\code\github\ePSproc\data\photoionization\n2_multiorb
- fileList :
- n2_3sg_0.1-50.1eV_A2.inp.out
- jobLabel :
- 3sg-1
- thres :
- None
Note here there are 3 polarization geometries set, \((x,y,z)\), which is the default case. The MF backend generates only MF results (no alignment dependence). For a quick intro to the MF and AF/LF calculations, see the ePSproc intro notes.
[9]:
# Params should already be OK... (set by demo script)
data.params
[9]:
| name | value | initial value | min | max | vary | expression |
|---|---|---|---|---|---|---|
| m_PU_SG_PU_1_n1_1_1 | 1.78461575 | 1.784615753610107 | 1.0000e-04 | 5.00000000 | True | |
| m_PU_SG_PU_1_1_n1_1 | 1.78461575 | 1.784615753610107 | 1.0000e-04 | 5.00000000 | False | m_PU_SG_PU_1_n1_1_1 |
| m_PU_SG_PU_3_n1_1_1 | 0.80290495 | 0.802904951323892 | 1.0000e-04 | 5.00000000 | True | |
| m_PU_SG_PU_3_1_n1_1 | 0.80290495 | 0.802904951323892 | 1.0000e-04 | 5.00000000 | False | m_PU_SG_PU_3_n1_1_1 |
| m_SU_SG_SU_1_0_0_1 | 2.68606212 | 2.686062120382649 | 1.0000e-04 | 5.00000000 | True | |
| m_SU_SG_SU_3_0_0_1 | 1.10915311 | 1.109153108617096 | 1.0000e-04 | 5.00000000 | True | |
| p_PU_SG_PU_1_n1_1_1 | -0.86104140 | -0.8610414024232179 | -3.14159265 | 3.14159265 | False | |
| p_PU_SG_PU_1_1_n1_1 | -0.86104140 | -0.8610414024232179 | -3.14159265 | 3.14159265 | False | p_PU_SG_PU_1_n1_1_1 |
| p_PU_SG_PU_3_n1_1_1 | -3.12044446 | -3.1204444620772467 | -3.14159265 | 3.14159265 | True | |
| p_PU_SG_PU_3_1_n1_1 | -3.12044446 | -3.1204444620772467 | -3.14159265 | 3.14159265 | False | p_PU_SG_PU_3_n1_1_1 |
| p_SU_SG_SU_1_0_0_1 | 2.61122920 | 2.611229196458127 | -3.14159265 | 3.14159265 | True | |
| p_SU_SG_SU_3_0_0_1 | -0.07867828 | -0.07867827542158025 | -3.14159265 | 3.14159265 | True |
Running a fit…
With the parameters and data set, everything is as per the previous demos - just call self.fit() to run a fit!
Statistics and outputs are handled by lmfit, which includes uncertainty estimates and correlations in the fitted parameters.
[10]:
# data.fit(fcn_kws = {'backend':ep.geomFunc.mfblmXprod}) # With backend by object
data.fit() # 21/08/22 With backend as set in data.fitOpts
[11]:
data.data.keys()
[11]:
dict_keys(['subset', 'orb6', 'orb5', 'ADM', 'pol', 'sim', 0])
Note the new result appears in the main data dict by fit number, here 0.
[12]:
# Check fit outputs
fitInd = 0
data.data[fitInd]['results']
[12]:
Fit Statistics
| fitting method | leastsq | |
| # function evals | 9 | |
| # data points | 75 | |
| # variables | 7 | |
| chi-square | 3.3126e-31 | |
| reduced chi-square | 4.8715e-33 | |
| Akaike info crit. | -5573.49196 | |
| Bayesian info crit. | -5557.26954 |
Variables
| name | value | standard error | relative error | initial value | min | max | vary | expression |
|---|---|---|---|---|---|---|---|---|
| m_PU_SG_PU_1_n1_1_1 | 1.78461575 | 2.3867e-09 | (0.00%) | 1.784615753610107 | 1.0000e-04 | 5.00000000 | True | |
| m_PU_SG_PU_1_1_n1_1 | 1.78461575 | 2.3867e-09 | (0.00%) | 1.784615753610107 | 1.0000e-04 | 5.00000000 | False | m_PU_SG_PU_1_n1_1_1 |
| m_PU_SG_PU_3_n1_1_1 | 0.80290495 | 1.0738e-09 | (0.00%) | 0.802904951323892 | 1.0000e-04 | 5.00000000 | True | |
| m_PU_SG_PU_3_1_n1_1 | 0.80290495 | 1.0738e-09 | (0.00%) | 0.802904951323892 | 1.0000e-04 | 5.00000000 | False | m_PU_SG_PU_3_n1_1_1 |
| m_SU_SG_SU_1_0_0_1 | 2.68606212 | 1.7419e-09 | (0.00%) | 2.686062120382649 | 1.0000e-04 | 5.00000000 | True | |
| m_SU_SG_SU_3_0_0_1 | 1.10915311 | 7.1926e-10 | (0.00%) | 1.109153108617096 | 1.0000e-04 | 5.00000000 | True | |
| p_PU_SG_PU_1_n1_1_1 | -0.86104140 | 0.00000000 | (0.00%) | -0.8610414024232179 | -3.14159265 | 3.14159265 | False | |
| p_PU_SG_PU_1_1_n1_1 | -0.86104140 | 0.00000000 | (0.00%) | -0.8610414024232179 | -3.14159265 | 3.14159265 | False | p_PU_SG_PU_1_n1_1_1 |
| p_PU_SG_PU_3_n1_1_1 | -3.12044446 | 1.0890e-16 | (0.00%) | -3.1204444620772467 | -3.14159265 | 3.14159265 | True | |
| p_PU_SG_PU_3_1_n1_1 | -3.12044446 | 1.0890e-16 | (0.00%) | -3.1204444620772467 | -3.14159265 | 3.14159265 | False | p_PU_SG_PU_3_n1_1_1 |
| p_SU_SG_SU_1_0_0_1 | 2.61122920 | 8.1419e-10 | (0.00%) | 2.611229196458127 | -3.14159265 | 3.14159265 | True | |
| p_SU_SG_SU_3_0_0_1 | -0.07867828 | 8.1419e-10 | (0.00%) | -0.07867827542158025 | -3.14159265 | 3.14159265 | True |
Correlations (unreported correlations are < 0.100)
| p_SU_SG_SU_1_0_0_1 | p_SU_SG_SU_3_0_0_1 | -1.0000 |
| m_PU_SG_PU_1_n1_1_1 | m_PU_SG_PU_3_n1_1_1 | -1.0000 |
| m_SU_SG_SU_1_0_0_1 | m_SU_SG_SU_3_0_0_1 | -1.0000 |
| m_PU_SG_PU_3_n1_1_1 | p_PU_SG_PU_3_n1_1_1 | -0.5828 |
| m_PU_SG_PU_1_n1_1_1 | p_PU_SG_PU_3_n1_1_1 | 0.5828 |
| m_SU_SG_SU_1_0_0_1 | p_SU_SG_SU_1_0_0_1 | 0.2621 |
| m_SU_SG_SU_1_0_0_1 | p_SU_SG_SU_3_0_0_1 | -0.2621 |
| m_SU_SG_SU_3_0_0_1 | p_SU_SG_SU_1_0_0_1 | -0.2621 |
| m_SU_SG_SU_3_0_0_1 | p_SU_SG_SU_3_0_0_1 | 0.2621 |
For this case, with correct inputs, correct outputs are quickly returned.
[13]:
# Quick plot with lmPlotFit - this defaults to the reference case plus the last fit
data.lmPlotFit(Etype='Labels')
Plotting data n2_3sg_0.1-50.1eV_A2.inp.out, pType=a, thres=0.01, with Seaborn
Plotting data (No filename), pType=a, thres=0.01, with Seaborn
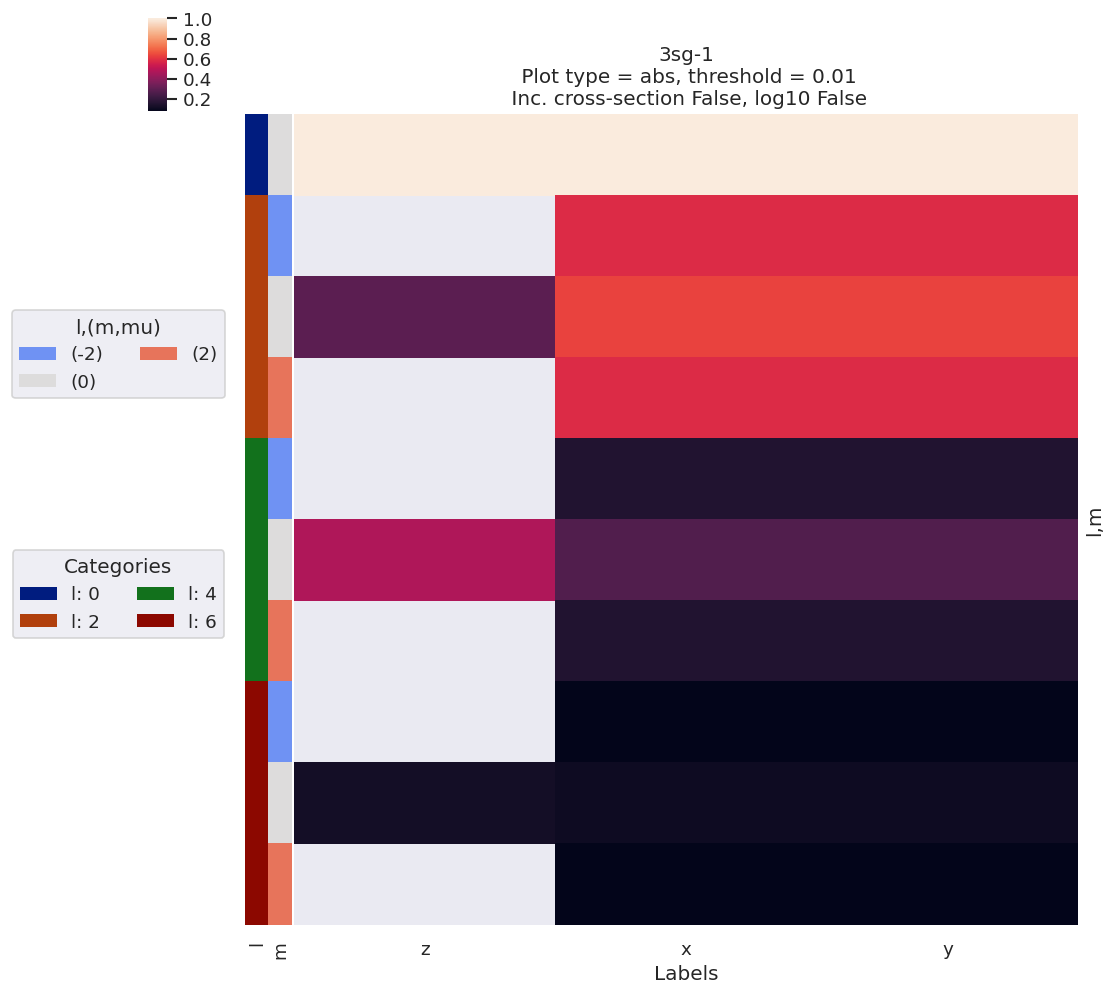
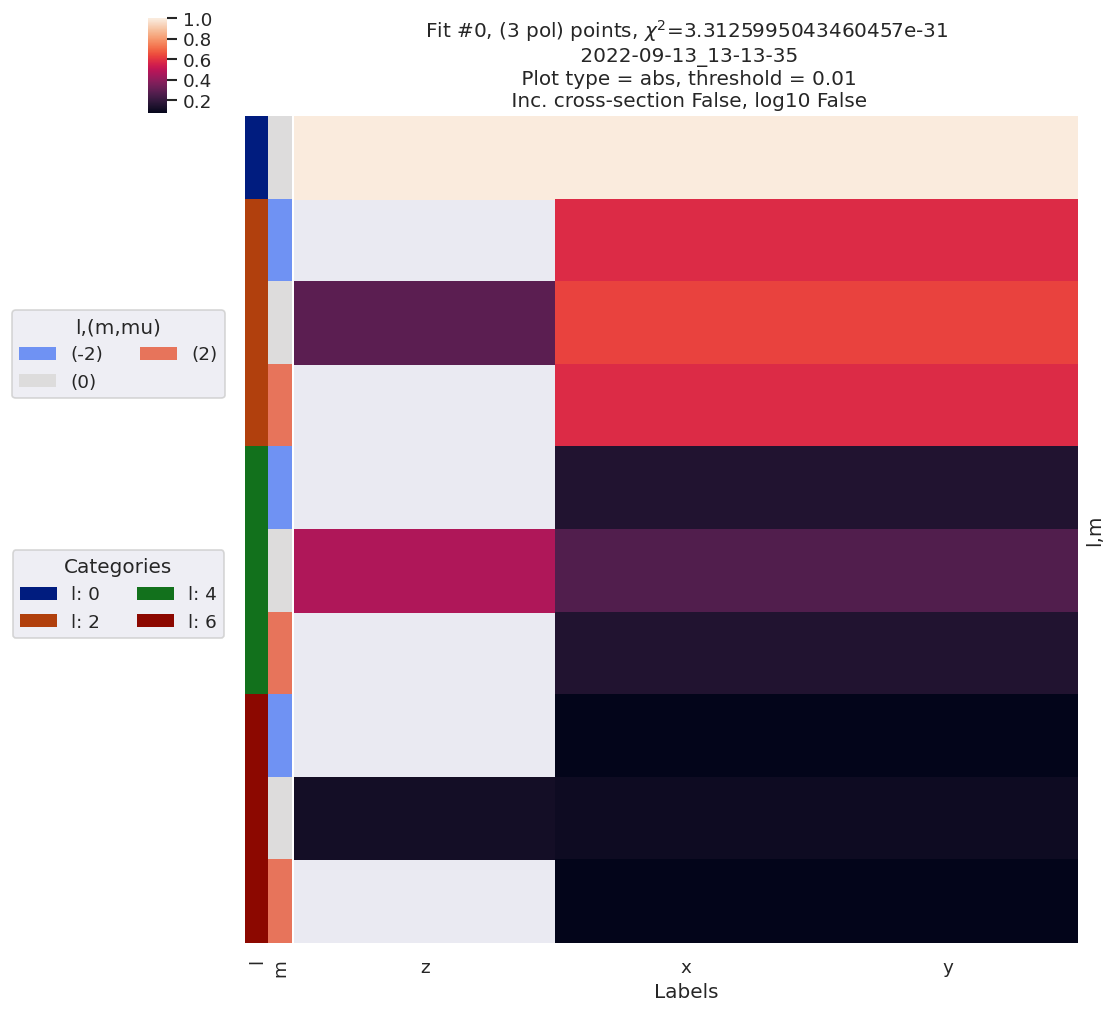
Fit with randomized params…
The basic case above is a useful sanity check for the fitting routine. Now we can test from unknown (randomized) inputs to confirm that the fitting is actually useful…
[14]:
# Basic randomize routine, [0,1] interval for input parameter set
data.randomizeParams()
# data.fit(fcn_kws = {'backend':ep.geomFunc.mfblmXprod}) # With backend by object
data.fit() # 21/08/22 With backend as set in data.fitOpts
data.lmPlotFit(Etype='Labels')
Plotting data n2_3sg_0.1-50.1eV_A2.inp.out, pType=a, thres=0.01, with Seaborn
Plotting data (No filename), pType=a, thres=0.01, with Seaborn
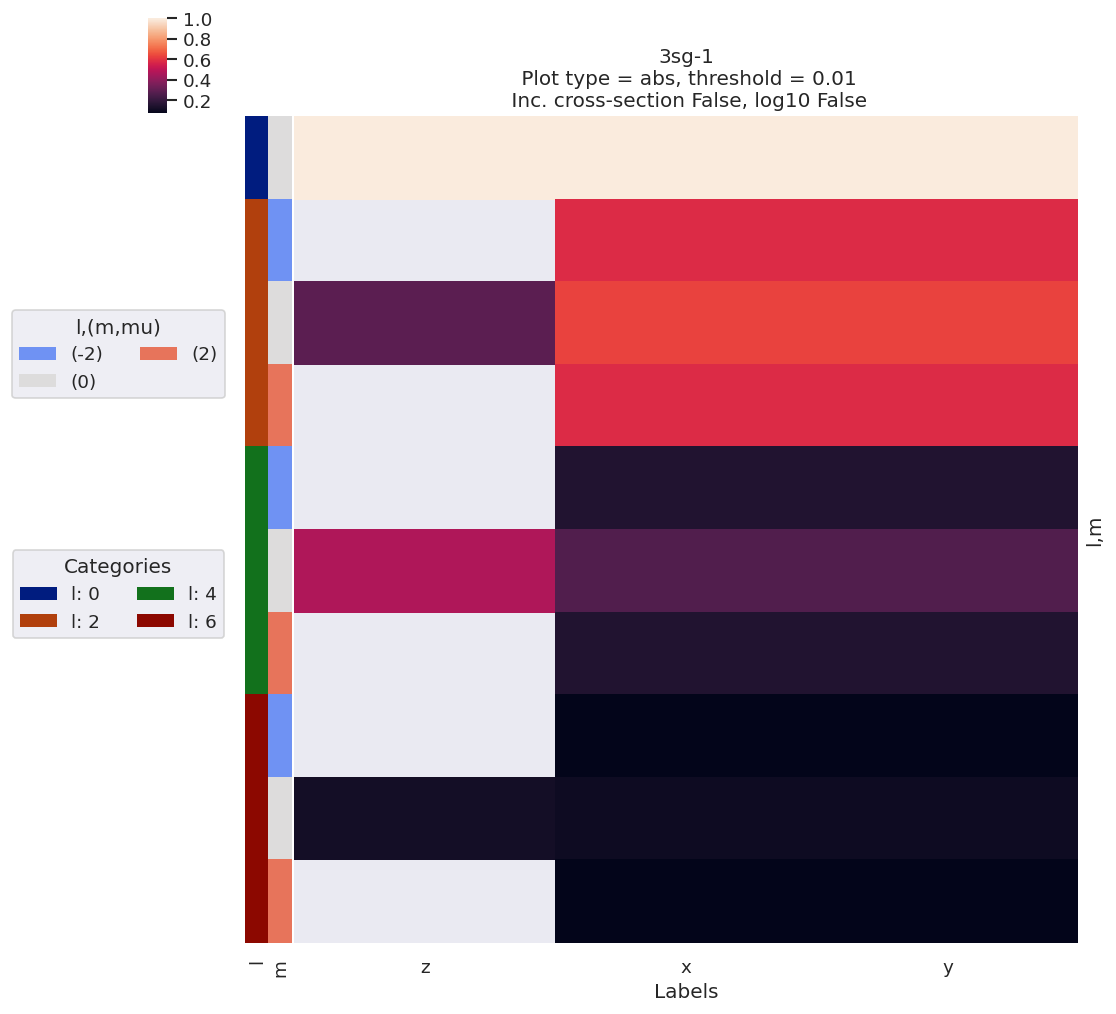
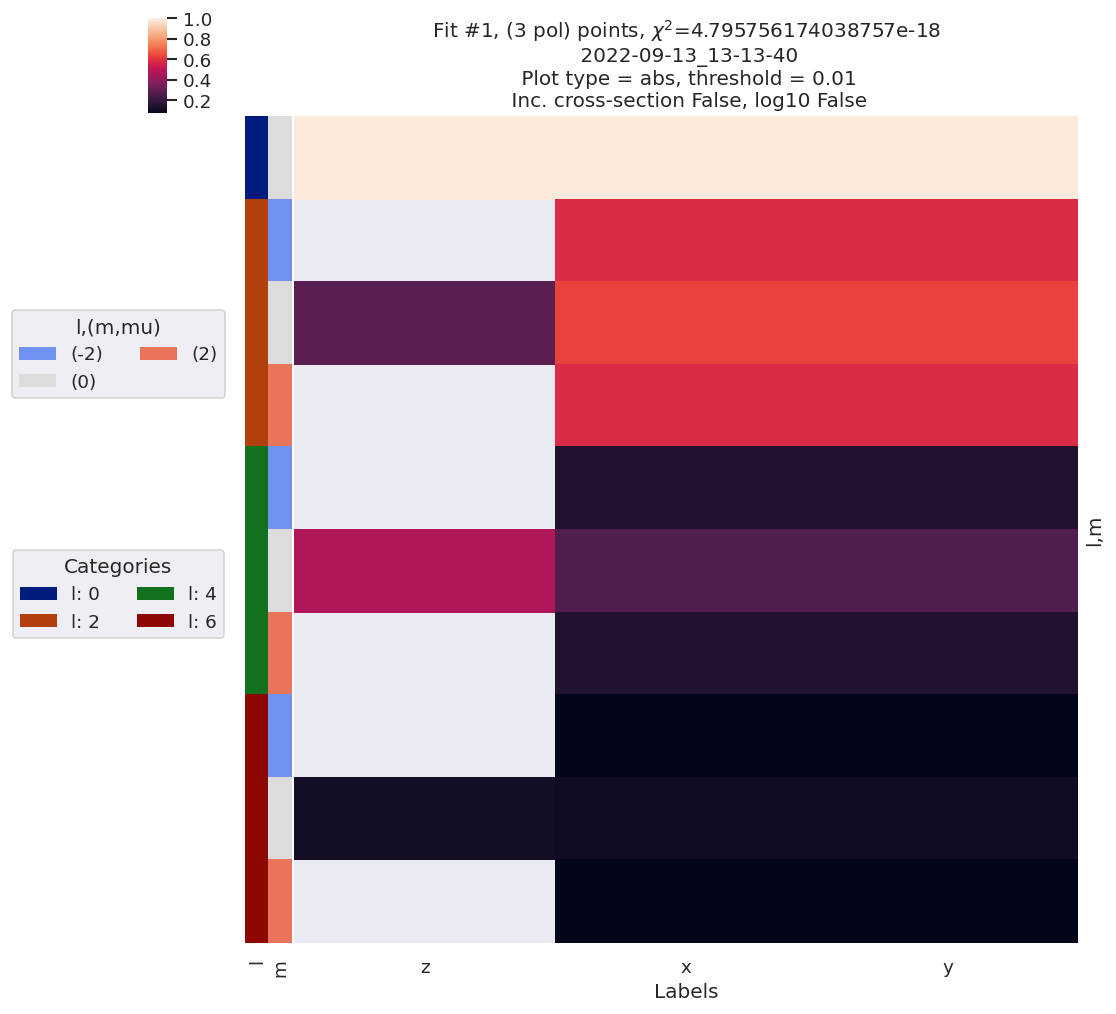
[15]:
# Check fit outputs
data.data[data.fitInd]['results']
[15]:
Fit Statistics
| fitting method | leastsq | |
| # function evals | 83 | |
| # data points | 75 | |
| # variables | 7 | |
| chi-square | 4.7958e-18 | |
| reduced chi-square | 7.0526e-20 | |
| Akaike info crit. | -3300.72163 | |
| Bayesian info crit. | -3284.49921 |
Variables
| name | value | standard error | relative error | initial value | min | max | vary | expression |
|---|---|---|---|---|---|---|---|---|
| m_PU_SG_PU_1_n1_1_1 | 0.08314518 | 4.9367e-07 | (0.00%) | 0.27981799849273714 | 1.0000e-04 | 5.00000000 | True | |
| m_PU_SG_PU_1_1_n1_1 | 0.08314518 | 0.00000000 | (0.00%) | 0.27981799849273714 | 1.0000e-04 | 5.00000000 | False | m_PU_SG_PU_1_n1_1_1 |
| m_PU_SG_PU_3_n1_1_1 | 0.03740731 | 2.2211e-07 | (0.00%) | 0.5537002130830051 | 1.0000e-04 | 5.00000000 | True | |
| m_PU_SG_PU_3_1_n1_1 | 0.03740731 | 0.00000000 | (0.00%) | 0.5537002130830051 | 1.0000e-04 | 5.00000000 | False | m_PU_SG_PU_3_n1_1_1 |
| m_SU_SG_SU_1_0_0_1 | 0.31810906 | nan | (nan%) | 0.6938920046815522 | 1.0000e-04 | 5.00000000 | True | |
| m_SU_SG_SU_3_0_0_1 | 0.13135648 | nan | (nan%) | 0.7601358965068807 | 1.0000e-04 | 5.00000000 | True | |
| p_PU_SG_PU_1_n1_1_1 | -0.86104140 | 0.00000000 | (0.00%) | -0.8610414024232179 | -3.14159265 | 3.14159265 | False | |
| p_PU_SG_PU_1_1_n1_1 | -0.86104140 | 0.00000000 | (0.00%) | -0.8610414024232179 | -3.14159265 | 3.14159265 | False | p_PU_SG_PU_1_n1_1_1 |
| p_PU_SG_PU_3_n1_1_1 | 1.39836166 | 4.9061e-10 | (0.00%) | 0.7549169336716605 | -3.14159265 | 3.14159265 | True | |
| p_PU_SG_PU_3_1_n1_1 | 1.39836166 | 0.00000000 | (0.00%) | 0.7549169336716605 | -3.14159265 | 3.14159265 | False | p_PU_SG_PU_3_n1_1_1 |
| p_SU_SG_SU_1_0_0_1 | -3.08684154 | nan | (nan%) | 0.5995617123509013 | -3.14159265 | 3.14159265 | True | |
| p_SU_SG_SU_3_0_0_1 | -0.39693408 | nan | (nan%) | 0.8621457643783 | -3.14159265 | 3.14159265 | True |
Correlations (unreported correlations are < 0.100)
| m_PU_SG_PU_1_n1_1_1 | m_PU_SG_PU_3_n1_1_1 | 1.0000 |
| m_PU_SG_PU_3_n1_1_1 | p_PU_SG_PU_3_n1_1_1 | 0.7263 |
| m_PU_SG_PU_1_n1_1_1 | p_PU_SG_PU_3_n1_1_1 | 0.7262 |
Note here that the \(\chi^2\) looks good, as do the MF-\(\beta_{LM}\), but the fitted parameters seem rather off from the reference case for the (m)agnitudes - this is because the input data was normalised by flux (i.e. \(\beta_{0,0}=1\)), so the fitting is only sensitive to relative magnitudes. The (p)hases, however, look good.
To get a better feel for this, run a batch of fits and look at the range of outputs more carefully…
Test batch run (multiple fits) with noisey data
Parallel run, see parallel execution for details.
In this test case, on an AMD Threadripper 2950X 16 core system, this took ~10 mins for 1000 fits.
[16]:
# Add noise with np.random.normal
# https://numpy.org/doc/stable/reference/random/generated/numpy.random.normal.html
# data.data['subset']['AFBLM']
import numpy as np
mu, sigma = 0, 0.05 # Up to approx 10% noise (+/- 0.05)
# creating a noise with the same dimension as the dataset (2,2)
noise = np.random.normal(mu, sigma, [data.data['subset']['AFBLM'].Labels.size, data.data['subset']['AFBLM'].l.size])
# data.BLMfitPlot()
# Set noise in Xarray & scale by l
import xarray as xr
noiseXR = xr.ones_like(data.data['subset']['AFBLM']) * noise
# data.data['subset']['AFBLM']['noise'] = ((data.data['subset']['AFBLM'].t, data.data['subset']['AFBLM'].l), noise)
# xr.where(noiseXR.l>0, noiseXR/noiseXR.l, noiseXR)
noiseXR = noiseXR.where(noiseXR.l<1, noiseXR/(noiseXR.l)) # Scale by L
data.data['subset']['AFBLM'] = data.data['subset']['AFBLM'] + noiseXR
data.data['subset']['AFBLM'] = data.data['subset']['AFBLM'].where(data.data['subset']['AFBLM'].m == 0, 0)
# data.BLMfitPlot(Etype='Labels', thres=None)
[17]:
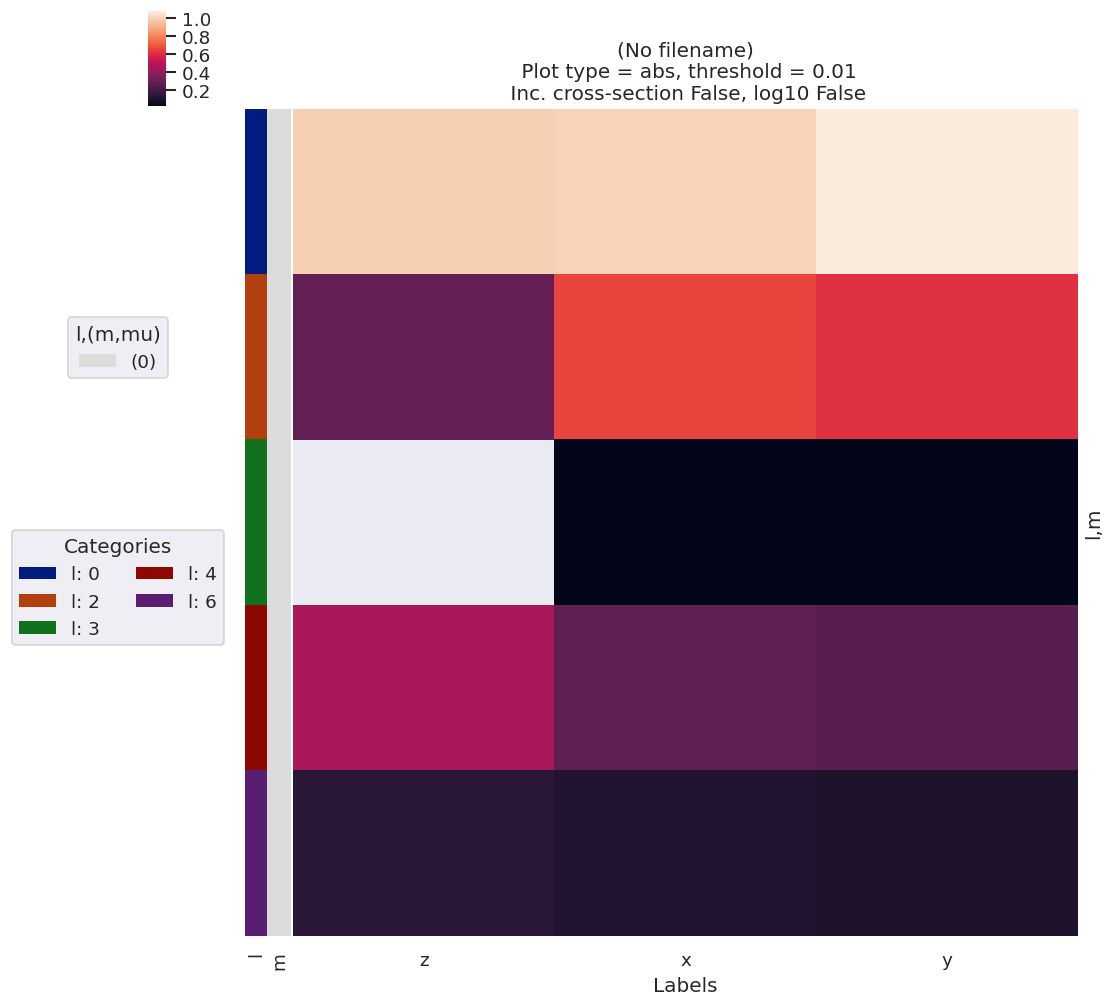
data.lmPlotFit(keys='subset',Etype='Labels')
Set dataType (No dataType)
Plotting data (No filename), pType=a, thres=0.01, with Seaborn
[18]:
data.multiFit(nRange = [0,1000])
100%|##############################################################################| 1000/1000 [13:33<00:00, 1.23it/s]
[19]:
# Data IO with self.writeFitData()
# This will default to the working dir and set a data-stamped file name if nothing is passed.
data.writeFitData(outStem='dataDump_100fitTests_MFBLM_multiFit_noise-3')
# data.writeFitData()
# Runs
# 1. dataDump_100fitTests_MFBLM_multiFit_noise-3_130922_10-42-09.pickle
# 2. dataDump_100fitTests_MFBLM_multiFit_noise-3_130922_13-27-15.pickle
[19]:
WindowsPath('D:/code/python/jupyterNotebooks/dev/PEMtk/fitting/MF_fit_dev_120822/dataDump_100fitTests_MFBLM_multiFit_noise-3_130922_13-27-15.pickle')
Data overview
For more on the data structures used here, see the setup & batch runs notebook.
For more on the analysis routines, see the fit fidelity and analysis routines notebooks.
[20]:
# Load data if required
# import pickle
# with open('dataDump_1000fitTests_MFBLM_multiFit_noise-1_220822.pickle', 'wb') as handle:
# pickle.dump(data.data, handle, protocol=pickle.HIGHEST_PROTOCOL)
# data.loadFitData('dataDump_1000fitTests_MFBLM_multiFit_noise-1_220822.pickle')
# data.loadFitData('dataDump_100fitTests_MFBLM_multiFit_noise-2_120922_19-34-54.pickle')
[21]:
# Run basic stats
# TODO: add more outputs here & tidy up output formatting.
data.analyseFits(keyDims='Labels')
*** Warning: found MultiIndex for DataFrame data.index - checkDims may have issues with Pandas MultiIndex, but will try anyway.
*** Warning: found MultiIndex for DataFrame data.index - checkDims may have issues with Pandas MultiIndex, but will try anyway.
[22]:
data.fitsSummary
[22]:
{'Fits': 999,
'Success': 999,
'Minima': {'chisqr': 1.34668471213695, 'redchi': 0.019804186943190444},
'Stats': {'chisqr': min 1.346685
mean 1.359765
median 1.346685
max 2.961047
std 0.074494
var 0.005549
Name: chisqr, dtype: float64,
'redchi': min 0.019804
mean 0.019997
median 0.019804
max 0.043545
std 0.001096
var 0.000001
Name: redchi, dtype: float64}}
[23]:
# Basic histogram of fit sets
# Note this defaults to Holoviews/Bokeh for plotting, which produces an interactive plot.
# Set backend = 'mpl' if Holoviews is not available.
data.fitHist(bins = 100)
[39]:
# Try zooming in on the good results - note in this example case the spread is small!
# data.fitHist(thres = 0.01992, bins = 100) # With threshold
# binRange = [0.0199115479, 0.0199115481]
# binRange = [0.01980418694, 0.019804187]
# binRange = [data.fitsSummary['Minima']['redchi'], data.fitsSummary['Minima']['redchi'] + data.fitsSummary['Minima']['redchi']*1e-8]
# Rough guess from variance?
binRange = [data.fitsSummary['Minima']['redchi'], data.fitsSummary['Minima']['redchi'] + data.fitsSummary['Stats']['redchi']['var']*1e-5]
data.fitHist(binRange = binRange, bins = 100, thres=binRange[1]) # With bins set, NOTE thres=binRange[1] sets X-lims here, now fixed in fitHist()
Mask selected 524 results (from 999).
[51]:
# Classify...
# data.classifyFits(bins = [0.01991154792, 0.019911548,20])
# data.classifyFits(bins = [0.01991154792, 0.01991154793,20]) # Slice best fit region
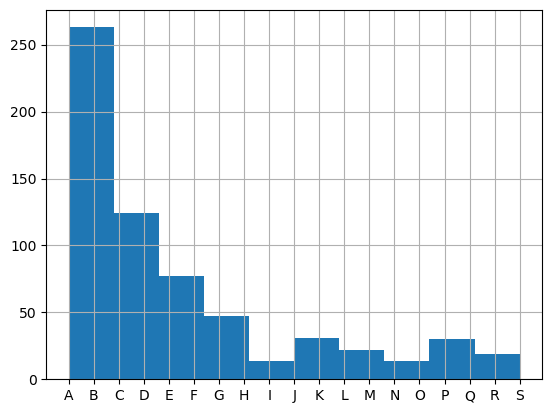
data.classifyFits(bins = [0.019804186942, 0.01980418697,20]) # Slice best fit region
# data.classifyFits(bins = [*binRange, 20])
| success | chisqr | redchi | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| count | unique | top | freq | count | unique | top | freq | count | unique | top | freq | |
| redchiGroup | ||||||||||||
| A | 42 | 1 | True | 42 | 42.0 | 42.0 | 1.346685 | 1.0 | 42.0 | 42.0 | 0.019804 | 1.0 |
| B | 221 | 1 | True | 221 | 221.0 | 221.0 | 1.346685 | 1.0 | 221.0 | 221.0 | 0.019804 | 1.0 |
| C | 76 | 1 | True | 76 | 76.0 | 76.0 | 1.346685 | 1.0 | 76.0 | 76.0 | 0.019804 | 1.0 |
| D | 48 | 1 | True | 48 | 48.0 | 48.0 | 1.346685 | 1.0 | 48.0 | 48.0 | 0.019804 | 1.0 |
| E | 53 | 1 | True | 53 | 53.0 | 53.0 | 1.346685 | 1.0 | 53.0 | 53.0 | 0.019804 | 1.0 |
| F | 24 | 1 | True | 24 | 24.0 | 24.0 | 1.346685 | 1.0 | 24.0 | 24.0 | 0.019804 | 1.0 |
| G | 18 | 1 | True | 18 | 18.0 | 18.0 | 1.346685 | 1.0 | 18.0 | 18.0 | 0.019804 | 1.0 |
| H | 29 | 1 | True | 29 | 29.0 | 29.0 | 1.346685 | 1.0 | 29.0 | 29.0 | 0.019804 | 1.0 |
| I | 14 | 1 | True | 14 | 14.0 | 14.0 | 1.346685 | 1.0 | 14.0 | 14.0 | 0.019804 | 1.0 |
| J | 13 | 1 | True | 13 | 13.0 | 13.0 | 1.346685 | 1.0 | 13.0 | 13.0 | 0.019804 | 1.0 |
| K | 18 | 1 | True | 18 | 18.0 | 18.0 | 1.346685 | 1.0 | 18.0 | 18.0 | 0.019804 | 1.0 |
| L | 14 | 1 | True | 14 | 14.0 | 14.0 | 1.346685 | 1.0 | 14.0 | 14.0 | 0.019804 | 1.0 |
| M | 8 | 1 | True | 8 | 8.0 | 8.0 | 1.346685 | 1.0 | 8.0 | 8.0 | 0.019804 | 1.0 |
| N | 10 | 1 | True | 10 | 10.0 | 10.0 | 1.346685 | 1.0 | 10.0 | 10.0 | 0.019804 | 1.0 |
| O | 4 | 1 | True | 4 | 4.0 | 4.0 | 1.346685 | 1.0 | 4.0 | 4.0 | 0.019804 | 1.0 |
| P | 18 | 1 | True | 18 | 18.0 | 18.0 | 1.346685 | 1.0 | 18.0 | 18.0 | 0.019804 | 1.0 |
| Q | 12 | 1 | True | 12 | 12.0 | 12.0 | 1.346685 | 1.0 | 12.0 | 12.0 | 0.019804 | 1.0 |
| R | 10 | 1 | True | 10 | 10.0 | 10.0 | 1.346685 | 1.0 | 10.0 | 10.0 | 0.019804 | 1.0 |
| S | 9 | 1 | True | 9 | 9.0 | 9.0 | 1.346685 | 1.0 | 9.0 | 9.0 | 0.019804 | 1.0 |
*** Warning: found MultiIndex for DataFrame data.index - checkDims may have issues with Pandas MultiIndex, but will try anyway.
*** Warning: found MultiIndex for DataFrame data.index - checkDims may have issues with Pandas MultiIndex, but will try anyway.
Examine trial parameter sets
[52]:
# Correct magnitudes (renorm) and phases (to ref)
data.phaseCorrection()
*** Warning: found MultiIndex for DataFrame data.index - checkDims may have issues with Pandas MultiIndex, but will try anyway.
*** Warning: found MultiIndex for DataFrame data.index - checkDims may have issues with Pandas MultiIndex, but will try anyway.
Set ref param = PU_SG_PU_1_1_n1_1
| Param | PU_SG_PU_1_1_n1_1 | PU_SG_PU_1_n1_1_1 | PU_SG_PU_3_1_n1_1 | PU_SG_PU_3_n1_1_1 | SU_SG_SU_1_0_0_1 | SU_SG_SU_3_0_0_1 | ||
|---|---|---|---|---|---|---|---|---|
| Fit | Type | redchiGroup | ||||||
| 0 | m | E | 1.771928 | 1.771928 | 0.830945 | 0.830945 | 4.986125 | 2.161857 |
| n | E | 0.290537 | 0.290537 | 0.136247 | 0.136247 | 0.817557 | 0.354472 | |
| p | E | -0.861041 | -0.861041 | 1.018645 | 1.018645 | 0.777926 | -2.888186 | |
| pc | E | -0.861041 | -0.861041 | 1.018645 | 1.018645 | 0.777926 | -2.888186 | |
| 2 | m | B | 4.695653 | 4.695653 | 2.201952 | 2.201952 | 4.956811 | 2.149144 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 996 | pc | B | -0.861041 | -0.861041 | -2.740728 | -2.740728 | -2.176238 | 0.440835 |
| 998 | m | I | 1.562246 | 1.562246 | 0.732626 | 0.732626 | 4.296351 | 1.862785 |
| n | I | 0.295854 | 0.295854 | 0.138743 | 0.138743 | 0.813631 | 0.352769 | |
| p | I | -0.861041 | -0.861041 | -2.740727 | -2.740727 | 3.123349 | -0.542763 | |
| pc | I | -0.861041 | -0.861041 | -2.740727 | -2.740727 | 3.123349 | -0.542763 |
2564 rows × 6 columns
[53]:
# Test best group, normalized magnitudes
plotGroup = 'A'
data.paramPlot(selectors={'Type':'n', 'redchiGroup':plotGroup}, hue = 'redchi', backend='hv', hvType='violin', remap = 'lmMap', hRound = 30) # New form with dict
*** Warning: found MultiIndex for DataFrame data.index - checkDims may have issues with Pandas MultiIndex, but will try anyway.
[54]:
# Test best group, corrected phases
data.paramPlot(selectors={'Type':'pc', 'redchiGroup':plotGroup}, hue = 'redchi', backend='hv', hvType='violin', remap = 'lmMap', hRound = 30) # New form with dict
*** Warning: found MultiIndex for DataFrame data.index - checkDims may have issues with Pandas MultiIndex, but will try anyway.
Based on these plots, in this case we can conclude:
There is some spread in the retrieved normalised magnitudes, but there are preferred regions of solution space, although the m=0 paramters are less well defined.
The sign of the phases was also undefined in this case (this is in line with the AF reconstruction procedure).
The relative phase between the \(\sigma_u\) continuum (m=0), and the \(\pi_u\) continuum is undefined in this case, which is due to a lack of cross-terms between continua in this case.
Overall, it seems like there is the expected level of convergence, and that the matrix elements were constrained as expect, with only relative magnitudes and phases obtained (per continuum) in this manner.
The values can be inspected a little more carefully by forcing the phases to abs values (i.e. \(0:\pi\) range), and comparing the normalized and phase-corrected values with the known inputs…
[55]:
data.phaseCorrection(absFlag=True, useRef = False) # Recompute phase corrections without reference shift & using 0:pi only
data.paramPlot(selectors={'Type':'pc', 'redchiGroup':plotGroup}, hue = 'redchi', backend='hv', hvType='violin', remap = 'lmMap', hRound = 30) # New form with dict
*** Warning: found MultiIndex for DataFrame data.index - checkDims may have issues with Pandas MultiIndex, but will try anyway.
*** Warning: found MultiIndex for DataFrame data.index - checkDims may have issues with Pandas MultiIndex, but will try anyway.
Set ref param = PU_SG_PU_1_1_n1_1
| Param | PU_SG_PU_1_1_n1_1 | PU_SG_PU_1_n1_1_1 | PU_SG_PU_3_1_n1_1 | PU_SG_PU_3_n1_1_1 | SU_SG_SU_1_0_0_1 | SU_SG_SU_3_0_0_1 | ||
|---|---|---|---|---|---|---|---|---|
| Fit | Type | redchiGroup | ||||||
| 0 | m | E | 1.771928 | 1.771928 | 0.830945 | 0.830945 | 4.986125 | 2.161857 |
| n | E | 0.290537 | 0.290537 | 0.136247 | 0.136247 | 0.817557 | 0.354472 | |
| p | E | -0.861041 | -0.861041 | 1.018645 | 1.018645 | 0.777926 | -2.888186 | |
| pc | E | 0.000000 | 0.000000 | 1.879686 | 1.879686 | 1.638968 | 2.027144 | |
| 2 | m | B | 4.695653 | 4.695653 | 2.201952 | 2.201952 | 4.956811 | 2.149144 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 996 | pc | B | 0.000000 | 0.000000 | 1.879686 | 1.879686 | 1.315196 | 1.301877 |
| 998 | m | I | 1.562246 | 1.562246 | 0.732626 | 0.732626 | 4.296351 | 1.862785 |
| n | I | 0.295854 | 0.295854 | 0.138743 | 0.138743 | 0.813631 | 0.352769 | |
| p | I | -0.861041 | -0.861041 | -2.740727 | -2.740727 | 3.123349 | -0.542763 | |
| pc | I | 0.000000 | 0.000000 | 1.879686 | 1.879686 | 2.298795 | 0.318278 |
2564 rows × 6 columns
*** Warning: found MultiIndex for DataFrame data.index - checkDims may have issues with Pandas MultiIndex, but will try anyway.
[56]:
# Get parameter statistics & compare with ref
# Specify subselection criteria with a dictionary of selectors (currently uses index values only) & report
data.paramsReport(inds = {'redchiGroup':plotGroup})
data.paramsCompare()
# Final comparison with formatting
import pandas as pd
# pd.options.display.precision = 5
pd.options.display.float_format = '{:.4f}'.format
data.paramsSummaryComp
*** Warning: found MultiIndex for DataFrame data.index - checkDims may have issues with Pandas MultiIndex, but will try anyway.
*** Warning: found MultiIndex for DataFrame data.index - checkDims may have issues with Pandas MultiIndex, but will try anyway.
*** Warning: found MultiIndex for DataFrame data.index - checkDims may have issues with Pandas MultiIndex, but will try anyway.
Set ref param = PU_SG_PU_1_1_n1_1
| Param | PU_SG_PU_1_1_n1_1 | PU_SG_PU_1_n1_1_1 | PU_SG_PU_3_1_n1_1 | PU_SG_PU_3_n1_1_1 | SU_SG_SU_1_0_0_1 | SU_SG_SU_3_0_0_1 | |
|---|---|---|---|---|---|---|---|
| Fit | Type | ||||||
| ref | m | 0.279818 | 0.279818 | 0.553700 | 0.553700 | 0.693892 | 0.760136 |
| n | 0.206901 | 0.206901 | 0.409412 | 0.409412 | 0.513072 | 0.562053 | |
| p | -0.861041 | -0.861041 | 0.754917 | 0.754917 | 0.599562 | 0.862146 | |
| pc | 0.000000 | 0.000000 | 1.615958 | 1.615958 | 1.460603 | 1.723187 |
C:\Users\femtolab\.conda\envs\ePSdev\lib\site-packages\IPython\core\interactiveshell.py:2886: PerformanceWarning: indexing past lexsort depth may impact performance.
return runner(coro)
[56]:
| Param | PU_SG_PU_1_1_n1_1 | PU_SG_PU_1_n1_1_1 | PU_SG_PU_3_1_n1_1 | PU_SG_PU_3_n1_1_1 | SU_SG_SU_1_0_0_1 | SU_SG_SU_3_0_0_1 | ||
|---|---|---|---|---|---|---|---|---|
| Type | Source | dType | ||||||
| m | mean | num | 2.9271 | 2.9271 | 1.3726 | 1.3726 | 2.4727 | 1.0721 |
| ref | num | 0.2798 | 0.2798 | 0.5537 | 0.5537 | 0.6939 | 0.7601 | |
| diff | % | 90.4405 | 90.4405 | 59.6610 | 59.6610 | 71.9377 | 29.0978 | |
| num | 2.6473 | 2.6473 | 0.8189 | 0.8189 | 1.7788 | 0.3120 | ||
| std | % | 60.7753 | 60.7753 | 60.7753 | 60.7753 | 78.9698 | 78.9698 | |
| num | 1.7790 | 1.7790 | 0.8342 | 0.8342 | 1.9527 | 0.8466 | ||
| diff/std | % | 148.8113 | 148.8113 | 98.1664 | 98.1664 | 91.0952 | 36.8467 | |
| n | mean | num | 0.4920 | 0.4920 | 0.2307 | 0.2307 | 0.4436 | 0.1924 |
| ref | num | 0.2069 | 0.2069 | 0.4094 | 0.4094 | 0.5131 | 0.5621 | |
| diff | % | 57.9463 | 57.9463 | 77.4567 | 77.4567 | 15.6510 | 192.2033 | |
| num | 0.2851 | 0.2851 | -0.1787 | -0.1787 | -0.0694 | -0.3697 | ||
| std | % | 35.7950 | 35.7950 | 35.7950 | 35.7950 | 66.7682 | 66.7683 | |
| num | 0.1761 | 0.1761 | 0.0826 | 0.0826 | 0.2962 | 0.1284 | ||
| diff/std | % | 161.8840 | 161.8840 | 216.3898 | 216.3898 | 23.4407 | 287.8663 | |
| p | mean | num | -0.8610 | -0.8610 | 0.0340 | 0.0340 | 0.6758 | -0.3214 |
| ref | num | -0.8610 | -0.8610 | 0.7549 | 0.7549 | 0.5996 | 0.8621 | |
| diff | % | 0.0000 | 0.0000 | 2117.2601 | 2117.2601 | 11.2857 | 368.2397 | |
| num | -0.0000 | -0.0000 | -0.7209 | -0.7209 | 0.0763 | -1.1836 | ||
| std | % | 0.0000 | 0.0000 | 4913.5282 | 4913.5282 | 314.8800 | 488.8465 | |
| num | 0.0000 | 0.0000 | 1.6729 | 1.6729 | 2.1281 | 1.5712 | ||
| diff/std | % | inf | inf | 43.0904 | 43.0904 | 3.5841 | 75.3283 | |
| pc | mean | num | 0.0000 | 0.0000 | 1.8797 | 1.8797 | 1.9511 | 1.2068 |
| ref | num | 0.0000 | 0.0000 | 1.6160 | 1.6160 | 1.4606 | 1.7232 | |
| diff | % | nan | nan | 14.0304 | 14.0304 | 25.1386 | 42.7879 | |
| num | 0.0000 | 0.0000 | 0.2637 | 0.2637 | 0.4905 | -0.5164 | ||
| std | % | nan | nan | 0.0000 | 0.0000 | 36.5002 | 64.9224 | |
| num | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.7121 | 0.7835 | ||
| diff/std | % | nan | nan | 521349978.3748 | 521349978.3748 | 68.8724 | 65.9061 |
These results follow from the conclusions above, with some additional caveats:
The fitted normalised magnitudes are excellent for \(\pi_u, l=1\), but less good for other cases (row type ‘n’, compared mean and ref values).
The fitted phases (‘pc’) are generally very good. Interestingly the \(\sigma_u\) mean phases look reasonable here, although this may just be a statistical artefact, since we expect a loss of relative phase information between the continua, as seen in the plots above. However, by setting these values as the reference case we should find a low spread of results (just uncorrelated with the \(\pi_u\) case).
[57]:
# Test sigma_u continua by setting different reference phase...
phaseCorrParams={'absFlag':True, 'useRef':False, 'refParam':'SU_SG_SU_1_0_0_1'}
# data.phaseCorrection(absFlag=True, useRef = False, refParam = 'SU_SG_SU_1_0_0_1') # Recompute phase corrections without reference shift & using 0:pi only
data.phaseCorrection(**phaseCorrParams)
data.paramPlot(selectors={'Type':'pc', 'redchiGroup':plotGroup}, hue = 'redchi', backend='hv', hvType='violin', remap = 'lmMap', hRound = 30) # New form with dict
# Specify subselection criteria with a dictionary of selectors (currently uses index values only) & report
data.paramsReport(inds = {'redchiGroup':plotGroup})
data.paramsCompare(phaseCorrParams=phaseCorrParams)
# Final comparison with formatting
import pandas as pd
# pd.options.display.precision = 5
pd.options.display.float_format = '{:.4f}'.format
data.paramsSummaryComp
*** Warning: found MultiIndex for DataFrame data.index - checkDims may have issues with Pandas MultiIndex, but will try anyway.
*** Warning: found MultiIndex for DataFrame data.index - checkDims may have issues with Pandas MultiIndex, but will try anyway.
Set ref param = SU_SG_SU_1_0_0_1
| Param | PU_SG_PU_1_1_n1_1 | PU_SG_PU_1_n1_1_1 | PU_SG_PU_3_1_n1_1 | PU_SG_PU_3_n1_1_1 | SU_SG_SU_1_0_0_1 | SU_SG_SU_3_0_0_1 | ||
|---|---|---|---|---|---|---|---|---|
| Fit | Type | redchiGroup | ||||||
| 0 | m | E | 1.7719 | 1.7719 | 0.8309 | 0.8309 | 4.9861 | 2.1619 |
| n | E | 0.2905 | 0.2905 | 0.1362 | 0.1362 | 0.8176 | 0.3545 | |
| p | E | -0.8610 | -0.8610 | 1.0186 | 1.0186 | 0.7779 | -2.8882 | |
| pc | E | 1.6390 | 1.6390 | 0.2407 | 0.2407 | 0.0000 | 2.6171 | |
| 2 | m | B | 4.6957 | 4.6957 | 2.2020 | 2.2020 | 4.9568 | 2.1491 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 996 | pc | B | 1.3152 | 1.3152 | 0.5645 | 0.5645 | 0.0000 | 2.6171 |
| 998 | m | I | 1.5622 | 1.5622 | 0.7326 | 0.7326 | 4.2964 | 1.8628 |
| n | I | 0.2959 | 0.2959 | 0.1387 | 0.1387 | 0.8136 | 0.3528 | |
| p | I | -0.8610 | -0.8610 | -2.7407 | -2.7407 | 3.1233 | -0.5428 | |
| pc | I | 2.2988 | 2.2988 | 0.4191 | 0.4191 | 0.0000 | 2.6171 |
2564 rows × 6 columns
*** Warning: found MultiIndex for DataFrame data.index - checkDims may have issues with Pandas MultiIndex, but will try anyway.
*** Warning: found MultiIndex for DataFrame data.index - checkDims may have issues with Pandas MultiIndex, but will try anyway.
*** Warning: found MultiIndex for DataFrame data.index - checkDims may have issues with Pandas MultiIndex, but will try anyway.
*** Warning: found MultiIndex for DataFrame data.index - checkDims may have issues with Pandas MultiIndex, but will try anyway.
Set ref param = SU_SG_SU_1_0_0_1
| Param | PU_SG_PU_1_1_n1_1 | PU_SG_PU_1_n1_1_1 | PU_SG_PU_3_1_n1_1 | PU_SG_PU_3_n1_1_1 | SU_SG_SU_1_0_0_1 | SU_SG_SU_3_0_0_1 | |
|---|---|---|---|---|---|---|---|
| Fit | Type | ||||||
| ref | m | 0.2798 | 0.2798 | 0.5537 | 0.5537 | 0.6939 | 0.7601 |
| n | 0.2069 | 0.2069 | 0.4094 | 0.4094 | 0.5131 | 0.5621 | |
| p | -0.8610 | -0.8610 | 0.7549 | 0.7549 | 0.5996 | 0.8621 | |
| pc | 1.4606 | 1.4606 | 0.1554 | 0.1554 | 0.0000 | 0.2626 |
C:\Users\femtolab\.conda\envs\ePSdev\lib\site-packages\IPython\core\interactiveshell.py:2886: PerformanceWarning: indexing past lexsort depth may impact performance.
return runner(coro)
[57]:
| Param | PU_SG_PU_1_1_n1_1 | PU_SG_PU_1_n1_1_1 | PU_SG_PU_3_1_n1_1 | PU_SG_PU_3_n1_1_1 | SU_SG_SU_1_0_0_1 | SU_SG_SU_3_0_0_1 | ||
|---|---|---|---|---|---|---|---|---|
| Type | Source | dType | ||||||
| m | mean | num | 2.9271 | 2.9271 | 1.3726 | 1.3726 | 2.4727 | 1.0721 |
| ref | num | 0.2798 | 0.2798 | 0.5537 | 0.5537 | 0.6939 | 0.7601 | |
| diff | % | 90.4405 | 90.4405 | 59.6610 | 59.6610 | 71.9377 | 29.0978 | |
| num | 2.6473 | 2.6473 | 0.8189 | 0.8189 | 1.7788 | 0.3120 | ||
| std | % | 60.7753 | 60.7753 | 60.7753 | 60.7753 | 78.9698 | 78.9698 | |
| num | 1.7790 | 1.7790 | 0.8342 | 0.8342 | 1.9527 | 0.8466 | ||
| diff/std | % | 148.8113 | 148.8113 | 98.1664 | 98.1664 | 91.0952 | 36.8467 | |
| n | mean | num | 0.4920 | 0.4920 | 0.2307 | 0.2307 | 0.4436 | 0.1924 |
| ref | num | 0.2069 | 0.2069 | 0.4094 | 0.4094 | 0.5131 | 0.5621 | |
| diff | % | 57.9463 | 57.9463 | 77.4567 | 77.4567 | 15.6510 | 192.2033 | |
| num | 0.2851 | 0.2851 | -0.1787 | -0.1787 | -0.0694 | -0.3697 | ||
| std | % | 35.7950 | 35.7950 | 35.7950 | 35.7950 | 66.7682 | 66.7683 | |
| num | 0.1761 | 0.1761 | 0.0826 | 0.0826 | 0.2962 | 0.1284 | ||
| diff/std | % | 161.8840 | 161.8840 | 216.3898 | 216.3898 | 23.4407 | 287.8663 | |
| p | mean | num | -0.8610 | -0.8610 | 0.0340 | 0.0340 | 0.6758 | -0.3214 |
| ref | num | -0.8610 | -0.8610 | 0.7549 | 0.7549 | 0.5996 | 0.8621 | |
| diff | % | 0.0000 | 0.0000 | 2117.2601 | 2117.2601 | 11.2857 | 368.2397 | |
| num | -0.0000 | -0.0000 | -0.7209 | -0.7209 | 0.0763 | -1.1836 | ||
| std | % | 0.0000 | 0.0000 | 4913.5282 | 4913.5282 | 314.8800 | 488.8465 | |
| num | 0.0000 | 0.0000 | 1.6729 | 1.6729 | 2.1281 | 1.5712 | ||
| diff/std | % | inf | inf | 43.0904 | 43.0904 | 3.5841 | 75.3283 | |
| pc | mean | num | 1.9511 | 1.9511 | 1.3527 | 1.3527 | 0.0000 | 2.6171 |
| ref | num | 1.4606 | 1.4606 | 0.1554 | 0.1554 | 0.0000 | 0.2626 | |
| diff | % | 25.1386 | 25.1386 | 88.5148 | 88.5148 | nan | 89.9665 | |
| num | 0.4905 | 0.4905 | 1.1973 | 1.1973 | 0.0000 | 2.3545 | ||
| std | % | 36.5002 | 36.5002 | 71.7079 | 71.7079 | nan | 0.0001 | |
| num | 0.7121 | 0.7121 | 0.9700 | 0.9700 | 0.0000 | 0.0000 | ||
| diff/std | % | 68.8724 | 68.8724 | 123.4380 | 123.4380 | nan | 107250322.9546 |
This again reflects a converged fit result, but a lack of correlation with the other continua. Additionally, the lack of sign on the phase has resulted in a phase-corrected value of 2.6, which is close to \(\pi-0.6\), i.e. \(\pi\) - (reference phase). In this case care must be taken in evaluation of the results! Adding a diagonal polarization case would fix most of these issue, by providing an observable sensitive to the phase relation between the continua.
Versions
[58]:
import scooby
scooby.Report(additional=['epsproc', 'pemtk', 'xarray', 'jupyter'])
[58]:
| Tue Sep 13 13:45:45 2022 Eastern Daylight Time | |||||
| OS | Windows | CPU(s) | 32 | Machine | AMD64 |
| Architecture | 64bit | RAM | 63.9 GB | Environment | Jupyter |
| Python 3.7.3 (default, Apr 24 2019, 15:29:51) [MSC v.1915 64 bit (AMD64)] | |||||
| epsproc | 1.3.2-dev | pemtk | 0.0.1 | xarray | 0.15.0 |
| jupyter | Version unknown | numpy | 1.18.1 | scipy | 1.3.0 |
| IPython | 7.12.0 | matplotlib | 3.3.1 | scooby | 0.5.6 |
| Intel(R) Math Kernel Library Version 2020.0.0 Product Build 20191125 for Intel(R) 64 architecture applications | |||||
[59]:
# Check current Git commit for local ePSproc version
from pathlib import Path
!git -C {Path(ep.__file__).parent} branch
!git -C {Path(ep.__file__).parent} log --format="%H" -n 1
* dev
master
numba-tests
815a839f3c8c4995532d6b5f87997b8ba2bb12eb
[60]:
# Check current remote commits
!git ls-remote --heads https://github.com/phockett/ePSproc
# !git ls-remote --heads git://github.com/phockett/epsman
92c661789a7d2927f2b53d7266f57de70b3834fa refs/heads/dependabot/pip/notes/envs/envs-versioned/mistune-2.0.3
fe1e9540c7b91fe571f60562acd31d8e489d491e refs/heads/dependabot/pip/notes/envs/envs-versioned/nbconvert-6.5.1
815a839f3c8c4995532d6b5f87997b8ba2bb12eb refs/heads/dev
1c0b8fd409648f07c85f4f20628b5ea7627e0c4e refs/heads/master
69cd89ce5bc0ad6d465a4bd8df6fba15d3fd1aee refs/heads/numba-tests
ea30878c842f09d525fbf39fa269fa2302a13b57 refs/heads/revert-9-master
[61]:
# Check current Git commit for local PEMtk version
import pemtk
from pathlib import Path
!git -C {Path(pemtk.__file__).parent} branch
!git -C {Path(pemtk.__file__).parent} log --format="%H" -n 1
* master
mfFittingDev
789b51ca06aa3b6b66123b27342be8b90f8d6a04
[62]:
# Check current remote commits
!git ls-remote --heads https://github.com/phockett/PEMtk
# !git ls-remote --heads git://github.com/phockett/epsman
aca9bc06bee4ab16dbe3d30a5b1e1b5604f975c5 refs/heads/master
3f4686dffdbb310f15692f978ba36d6a3d15e8d3 refs/heads/mfFittingDev