Figure and data processing for Topical Review: Extracting Molecular Frame Photoionization Dynamics from Experimental Data
18/07/22
This document illustrates the use of the Photoelectron Metrology Toolkit (PEMtk) for analysis of matrix element fitting results, plus some additional plotting routines. Results and figures are as reported in the manuscript Topical Review: Extracting Molecular Frame Photoionization Dynamics from Experimental Data [1], available via Authorea.
Full data and code (current version) available via Figshare, DOI: 10.6084/m9.figshare.20293782.
For general info, see the PEMtk analysis routines docs.
v29/07/22: tidied up, fixed fig panel labels.
v26/07/22: updated plots now in place, just needs a tidy up.
v18/07/22: adapted from original processing notebooks 23/04/22, tidied up for distribution, plus some PEMtk updates incorporated.
For MFPAD surface plots see the notebook MFPAD_replotting_from_file_190722-dist.ipynb.
Setup
Load fit & analysis class and import data (currently a bit messy).
[1]:
saveTables = False
saveFigs = True # Set to save figs to file
exportData = False # Save processed data to file
[2]:
# Import & set paths
import pemtk
from pemtk.fit.fitClass import pemtkFit
from pathlib import Path
# Path for demo script
demoPath = Path(pemtk.__file__).parent.parent/Path('demos','fitting')
# Load sample dataset
dataPath = demoPath
# Basic test data - simulated results with no noise, 100 fits
# dataFile = 'dataDump_100fitTests_10t_randPhase_130621.pickle'
# Noisey test data - simulated results with noise, 1000 fits
dataFile = 'dataDump_1000fitTests_multiFit_noise_051021.pickle' # Full class Pickle
# dataFile = 'dataDump_130722_12-21-17.pdHDF' # Pandas hdf version
# 13/07/22 - with updated IO routines
data = pemtkFit()
data.loadFitData(dataFile,dataPath)
*** ePSproc installation not found, setting for local copy.
* Setting plotter defaults with epsproc.basicPlotters.setPlotters(). Run directly to modify, or change options in local env.
* Set Holoviews with bokeh.
C:\Users\femtolab\.conda\envs\ePSdev\lib\site-packages\xyzpy\plot\xyz_cmaps.py:6: MatplotlibDeprecationWarning:
The revcmap function was deprecated in Matplotlib 3.2 and will be removed two minor releases later. Use Colormap.reversed() instead.
return LinearSegmentedColormap(name, cm.revcmap(cmap._segmentdata))
Read data from d:\code\github\pemtk\demos\fitting\dataDump_1000fitTests_multiFit_noise_051021.pickle with pickle.
Additional figure handling options
[3]:
# Some additional default plot settings
# TODO: this is already run at class init, but out of notebook scope? Should fix.
import epsproc as ep # For some low-level routines
from epsproc.plot import hvPlotters # Additional plotting code
hvPlotters.setPlotters(snsStyle='white')
# hvPlotters.sns.set_style("white") # No grid case (for maps)
hv = hvPlotters.hv # Set usual alias
# General size & range unless overridden
figSize = [700,300]
tRange = [3.8, 5.2] # Set for axis slice, also for cmapping lims
# Update defaults
hvPlotters.setPlotters(width = figSize[0], height = figSize[1], snsStyle = 'white')
# Turn off Bokeh warnings
import warnings
# warnings.filterwarnings("ignore", message="when aspect and width/height is set, the bokeh backend")
warnings.simplefilter('ignore')
* Set Holoviews with bokeh.
* Set Holoviews with bokeh.
[4]:
# Pandas table display settings
import pandas as pd
# pd.options.display.precision = 5
pd.options.display.float_format = '{:.3f}'.format
[5]:
# Optional Matplotlib & set timeString (or will be set by hvSave in most cases)
# import matplotlib.pyplot as plt
# from datetime import datetime as dt
# timeString = dt.now()
Preprocessing
[6]:
# Run basic stats
data.analyseFits()
Pandas reference table not set, missing self.params data.
*** Warning: found MultiIndex for DataFrame data.index - checkDims may have issues with Pandas MultiIndex, but will try anyway.
*** Warning: found MultiIndex for DataFrame data.index - checkDims may have issues with Pandas MultiIndex, but will try anyway.
{ 'Fits': 1000,
'Minima': {'chisqr': 0.04215746268613921, 'redchi': 0.00022911664503336526},
'Success': 992}
Data conversion & export
Convert Pickled class data > more robust formats for distribution!
Note - this is aggregate data or specific fit data only (saves Pandas Dataframes or Xarray currently), but should be sufficient to recover results if Pickle file IO breaks.
[7]:
# Write single key:value pairs
# data.writeFitData(outStem = 'dataDump_1000fitTests_multiFit_noise_051021', fType='pdHDF')
[8]:
# Write agg datasets.
if exportData:
data.processedToHDF5(dataPath = dataPath, outStem = 'dataDump_1000fitTests_multiFit_noise_051021', timeStamp=False)
Also export current env for transparency
[9]:
condaEnv = 'epsdev'
[10]:
!conda run -n {condaEnv} conda env export --no-builds > {Path(dataPath,'epsdev_051021.yml').as_posix()}
C:\Users\femtolab\AppData\Roaming\Python\Python37\site-packages\requests\__init__.py:91: RequestsDependencyWarning: urllib3 (1.25.8) or chardet (3.0.4) doesn't match a supported version!
RequestsDependencyWarning)
C:\Users\femtolab\AppData\Roaming\Python\Python37\site-packages\requests\__init__.py:91: RequestsDependencyWarning: urllib3 (1.25.8) or chardet (3.0.4) doesn't match a supported version!
RequestsDependencyWarning)
[11]:
!conda run -n {condaEnv} pip list --format=freeze > {Path(dataPath,'epsdev_051021_requirements.txt').as_posix()}
[12]:
# Set for fig saving
data.job['fileIn'] = Path(dataFile)
data.job['fileBase'] = Path(data.job['fileBase'], 'figs_290722')
Input data and alignment plots
Plot ADMs and \(\beta_{LM}\) inputs to fit, then stack to multi-panel plot.
[13]:
# For ADMs
# Bespoke handling, see https://epsproc.readthedocs.io/en/dev/tests/hvPlotters_fn_tests_150720_v250122.html#curvePlot()-for-general-Holoviews-plotting
key = 'subset'
dataType='ADM'
daPlot = ep.matEleSelector(data.data[key][dataType], thres=1e-2, dims = 't', sq = True).squeeze()
daPlot = ep.plotTypeSelector(daPlot, pType = 'r')
from holoviews import opts, dim
# Plot with widgets to handle dims - note this may require .unstack() for multindex objects
hvObj = hvPlotters.curvePlot(daPlot.rename(r'$\beta_{LM}$').unstack(), kdims='t', renderPlot=False).opts(line_dash='dashed')
hvObjP = hvObj * daPlot.rename(r'$\beta_{LM}$').unstack().hvplot.scatter(x='t', marker = 'x', size = 100) # Add points
key = 'ADM'
dataType='ADM'
daPlot = ep.matEleSelector(data.data[key][dataType], thres=1e-2, dims = 't', sq = True).squeeze()
daPlot = ep.plotTypeSelector(daPlot, pType = 'r')
# Plot with widgets to handle dims - note this may require .unstack() for multindex objects
hvObj2 = hvPlotters.curvePlot(daPlot.rename(r'$\beta_{LM}$').unstack(), kdims='t', renderPlot=False)
# Overlay plot
hvADMPlot = (hvObjP * hvObj2).overlay(['K','Q','S'])
[14]:
# Add BLMs...
key = 'subset'
dataType='AFBLM'
# blmLabel = r'$\beta_{LM}$' # Doens't work in Bokeh, unless v>2.4, see https://github.com/bokeh/bokeh/issues/6031
blmLabel = 'βlm (unnorm)' # No latex
daPlot = ep.matEleSelector(data.data[key][dataType], thres=1e-2, dims = 't', sq = True).squeeze()
daPlot = ep.plotTypeSelector(daPlot, pType = 'r')
from holoviews import opts, dim
# Plot with widgets to handle dims - note this may require .unstack() for multindex objects
hvObj = hvPlotters.curvePlot(daPlot.rename(blmLabel).unstack(), kdims='t', renderPlot=False).opts(line_dash='dashed')
hvObjP = hvObj * daPlot.rename(blmLabel).unstack().hvplot.scatter(x='t', marker = 'x', size = 100) # Add points
key = 'sim'
# dataType='ADM'
daPlot = ep.matEleSelector(data.data[key][dataType], thres=1e-2, dims = 't', sq = True).squeeze()
daPlot = ep.plotTypeSelector(daPlot, pType = 'r')
# Plot with widgets to handle dims - note this may require .unstack() for multindex objects
hvObj2 = hvPlotters.curvePlot(daPlot.rename(blmLabel).unstack(), kdims='t', renderPlot=False)
hvBLMPlot = (hvObjP * hvObj2).overlay(['l','m'])
[15]:
# Expand ADMs to P(theta)
data.data['ADM']['ADM'].attrs['jobLabel'] = 'ADMs'
# Old version
# ADMdistFull = data.padPlot(keys = 'ADM', dataType='ADM', Etype = 't', pType='r', pStyle='grid', reducePhi='sum', returnFlag = True, sumDims = {})
# Updated 23/04/22
data.padPlot(keys = 'ADM', dataType='ADM', Etype = 't', pType='r', pStyle='grid', reducePhi='sum', returnFlag = True, sumDims = {})
ADMdistFull = data.data['ADM']['plots']['ADM']['pData']
Using default sph betas.
Plotting from self.data[ADM][ADM], facetDims=['Labels', 't'], pType=r with backend=mpl.
Grid plot: ADMs, dataType: ADM, plotType: r
Set plot to self.data['ADM']['plots']['ADM']['grid']
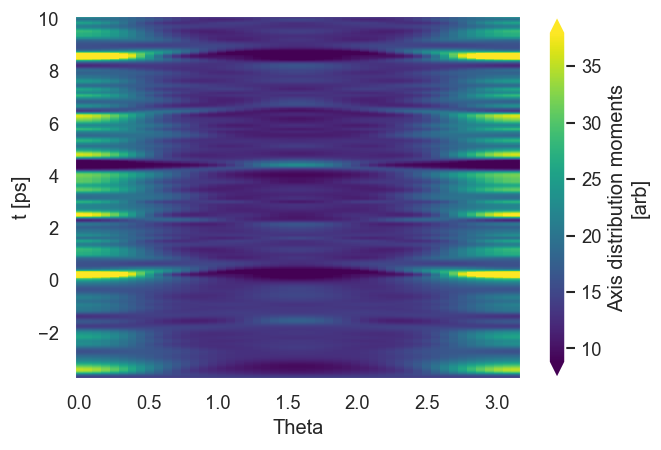
[16]:
# Expand BLMs to I(theta)
key = 'sim'
dataType = 'AFBLM'
# data.data[key][dataType].attrs['jobLabel'] = 'ADMs'
# Old version
# It = data.padPlot(keys = key, dataType=dataType, Etype = 't', pType='r', pStyle='grid', reducePhi='sum', returnFlag = True, sumDims = {}, selDims = {'Labels':'A'})
# Updated 23/04/22
data.padPlot(keys = key, dataType=dataType, Etype = 't', pType='r', pStyle='grid', reducePhi='sum', returnFlag = True, sumDims = {}, selDims = {'Labels':'A'})
It = data.data['sim']['plots']['AFBLM']['pData']
Using default sph betas.
Plotting from self.data[sim][AFBLM], facetDims=['Labels', 't'], pType=r with backend=mpl.
Grid plot: 3sg-1, dataType: AFBLM, plotType: r
Set plot to self.data['sim']['plots']['AFBLM']['grid']
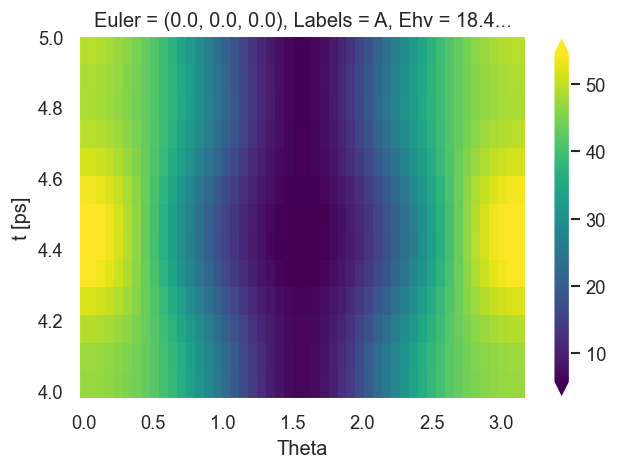
[17]:
# 4up plot layout - ADMs & BLMs + theta,E maps
# Can't suppress warnings here?
# %%capture --no-stderr
# warnings.simplefilter('ignore')
# figSize = [700,300]
# tRange = [3.8, 5.2] # Set for axis slice, also for cmapping lims
# hvPlotters.sns.set_style("white") # No grid case (for maps) - Holoviews picking this up...?
ADMdistPlot = ADMdistFull.sel({'t':slice(tRange[0],tRange[1])}) # Subselect this dataset first, otherwise it's all output to file and it is ~12Mb (6.1Mb limit on Authorea)
layout2 = (hvADMPlot.opts(frame_width = figSize[0], frame_height = figSize[1], xaxis=None, ylim=(-0.4,1.1), title='(a) ADMs') +
(ADMdistPlot/ADMdistPlot.max()).rename('v').squeeze().hvplot.heatmap(x='t',y='Theta',ylabel='θ',C='v', xaxis=None, cmap='viridis', frame_width = figSize[0], frame_height = figSize[1], title=r'(b) P(θ,t)').redim.range(v=(0, 1.0)) +
hvBLMPlot.opts(frame_width = figSize[0], frame_height = figSize[1], xaxis=None, title=r'(c) βlm(t)') +
(It/It.max()).rename('v').squeeze().hvplot.heatmap(x='t',y='Theta',ylabel='θ',C='v', cmap='viridis', frame_width = figSize[0], frame_height = figSize[1], title=r'(d) I(θ,t)')).redim(t=hv.Dimension('t', range=(tRange[0], tRange[1]))).cols(1)
pType = 'data4up'
data.data['plots'] = {} # Init plots dict
data.data['plots'][pType] = layout2
if saveFigs:
data.hvSave(pTypes = pType)
# Show plot
layout2
Saving self.data[plots][data4up] to D:\code\python\jupyterNotebooks\dev\PEMtk\fitting\multiFit_tests_and_parallel\figs_290722\dataDump_1000fitTests_multiFit_noise_051021_data4up_290722_11-25-39.png
WARNING:param.HeatMapPlot13372: Due to internal constraints, when aspect and width/height is set, the bokeh backend uses those values as frame_width/frame_height instead. This ensures the aspect is respected, but means that the plot might be slightly larger than anticipated. Set the frame_width/frame_height explicitly to suppress this warning.
WARNING:param.HeatMapPlot13372: aspect value was ignored because absolute width and height values were provided. Either supply explicit frame_width and frame_height to achieve desired aspect OR supply a combination of width or height and an aspect value.
WARNING:param.HeatMapPlot14283: Due to internal constraints, when aspect and width/height is set, the bokeh backend uses those values as frame_width/frame_height instead. This ensures the aspect is respected, but means that the plot might be slightly larger than anticipated. Set the frame_width/frame_height explicitly to suppress this warning.
WARNING:param.HeatMapPlot14283: aspect value was ignored because absolute width and height values were provided. Either supply explicit frame_width and frame_height to achieve desired aspect OR supply a combination of width or height and an aspect value.
Saving self.data[plots][data4up] to D:\code\python\jupyterNotebooks\dev\PEMtk\fitting\multiFit_tests_and_parallel\figs_290722\dataDump_1000fitTests_multiFit_noise_051021_data4up_290722_11-25-50.html
WARNING:param.HeatMapPlot18402: Due to internal constraints, when aspect and width/height is set, the bokeh backend uses those values as frame_width/frame_height instead. This ensures the aspect is respected, but means that the plot might be slightly larger than anticipated. Set the frame_width/frame_height explicitly to suppress this warning.
WARNING:param.HeatMapPlot18402: aspect value was ignored because absolute width and height values were provided. Either supply explicit frame_width and frame_height to achieve desired aspect OR supply a combination of width or height and an aspect value.
WARNING:param.HeatMapPlot19313: Due to internal constraints, when aspect and width/height is set, the bokeh backend uses those values as frame_width/frame_height instead. This ensures the aspect is respected, but means that the plot might be slightly larger than anticipated. Set the frame_width/frame_height explicitly to suppress this warning.
WARNING:param.HeatMapPlot19313: aspect value was ignored because absolute width and height values were provided. Either supply explicit frame_width and frame_height to achieve desired aspect OR supply a combination of width or height and an aspect value.
WARNING:param.HeatMapPlot23525: Due to internal constraints, when aspect and width/height is set, the bokeh backend uses those values as frame_width/frame_height instead. This ensures the aspect is respected, but means that the plot might be slightly larger than anticipated. Set the frame_width/frame_height explicitly to suppress this warning.
WARNING:param.HeatMapPlot23525: aspect value was ignored because absolute width and height values were provided. Either supply explicit frame_width and frame_height to achieve desired aspect OR supply a combination of width or height and an aspect value.
WARNING:param.HeatMapPlot24436: Due to internal constraints, when aspect and width/height is set, the bokeh backend uses those values as frame_width/frame_height instead. This ensures the aspect is respected, but means that the plot might be slightly larger than anticipated. Set the frame_width/frame_height explicitly to suppress this warning.
WARNING:param.HeatMapPlot24436: aspect value was ignored because absolute width and height values were provided. Either supply explicit frame_width and frame_height to achieve desired aspect OR supply a combination of width or height and an aspect value.
[17]:
Classifiers & best fits
Fitting histogram
[18]:
# With threshold set & fine binning.
# See fitHist() docs for more options.
data.fitHist(thres = 2.4e-4, bins = 100)
# data.fitHist(bins=20, binRange = [2.3e-4, 2.4e-4]) # Example with a range set
Mask selected 562 results (from 1000).
[20]:
if saveFigs:
data.hvSave(pTypes = 'fitHistPlot') # , outStem = 'test')
Saving self.data[plots][fitHistPlot] to D:\code\python\jupyterNotebooks\dev\PEMtk\fitting\multiFit_tests_and_parallel\figs_290722\dataDump_1000fitTests_multiFit_noise_051021_fitHistPlot_290722_11-26-28.png
Saving self.data[plots][fitHistPlot] to D:\code\python\jupyterNotebooks\dev\PEMtk\fitting\multiFit_tests_and_parallel\figs_290722\dataDump_1000fitTests_multiFit_noise_051021_fitHistPlot_290722_11-26-30.html
Rebin data by reduced \(\chi^2\) (small range)
[21]:
# Again, more control can be obtained by specifiying the desired binning.
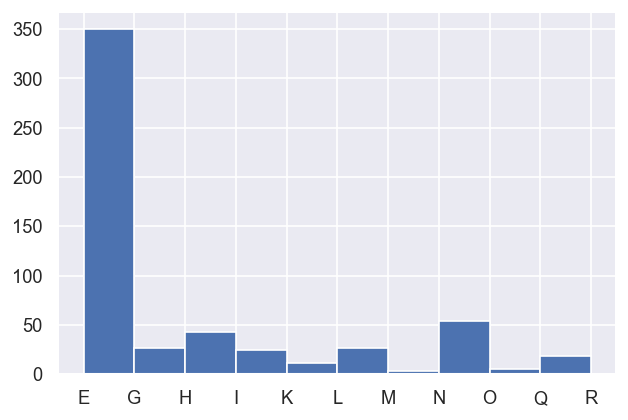
data.classifyFits(bins = [2.26e-4, 2.4e-4,20])
| success | chisqr | redchi | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| count | unique | top | freq | count | unique | top | freq | count | unique | top | freq | |
| redchiGroup | ||||||||||||
| A | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN |
| B | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN |
| C | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN |
| D | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN |
| E | 350 | 1 | True | 350 | 350.000 | 350.000 | 0.042 | 1.000 | 350.000 | 350.000 | 0.000 | 1.000 |
| F | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN |
| G | 27 | 1 | True | 27 | 27.000 | 27.000 | 0.043 | 1.000 | 27.000 | 27.000 | 0.000 | 1.000 |
| H | 43 | 1 | True | 43 | 43.000 | 43.000 | 0.043 | 1.000 | 43.000 | 43.000 | 0.000 | 1.000 |
| I | 24 | 1 | True | 24 | 24.000 | 24.000 | 0.043 | 1.000 | 24.000 | 24.000 | 0.000 | 1.000 |
| J | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN |
| K | 11 | 1 | True | 11 | 11.000 | 11.000 | 0.043 | 1.000 | 11.000 | 11.000 | 0.000 | 1.000 |
| L | 27 | 1 | True | 27 | 27.000 | 27.000 | 0.043 | 1.000 | 27.000 | 27.000 | 0.000 | 1.000 |
| M | 3 | 2 | True | 2 | 3.000 | 3.000 | 0.043 | 1.000 | 3.000 | 3.000 | 0.000 | 1.000 |
| N | 54 | 1 | True | 54 | 54.000 | 54.000 | 0.043 | 1.000 | 54.000 | 54.000 | 0.000 | 1.000 |
| O | 5 | 1 | True | 5 | 5.000 | 5.000 | 0.044 | 1.000 | 5.000 | 5.000 | 0.000 | 1.000 |
| P | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN |
| Q | 15 | 2 | True | 14 | 15.000 | 15.000 | 0.044 | 1.000 | 15.000 | 15.000 | 0.000 | 1.000 |
| R | 3 | 1 | True | 3 | 3.000 | 3.000 | 0.044 | 1.000 | 3.000 | 3.000 | 0.000 | 1.000 |
| S | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN |
*** Warning: found MultiIndex for DataFrame data.index - checkDims may have issues with Pandas MultiIndex, but will try anyway.
*** Warning: found MultiIndex for DataFrame data.index - checkDims may have issues with Pandas MultiIndex, but will try anyway.
[22]:
data.setMatEFit() # In current dataset data.lmmu not present initially? Run this to generate
# data.data['plots']['paramData'].replace({'Param':data.lmmu['lmMap']})
Set 6 complex matrix elements to 12 fitting params, see self.params for details.
Auto-setting parameters.
| name | value | initial value | min | max | vary | expression |
|---|---|---|---|---|---|---|
| m_PU_SG_PU_1_n1_1_1 | 1.78461575 | 1.784615753610107 | 1.0000e-04 | 5.00000000 | True | |
| m_PU_SG_PU_1_1_n1_1 | 1.78461575 | 1.784615753610107 | 1.0000e-04 | 5.00000000 | False | m_PU_SG_PU_1_n1_1_1 |
| m_PU_SG_PU_3_n1_1_1 | 0.80290495 | 0.802904951323892 | 1.0000e-04 | 5.00000000 | True | |
| m_PU_SG_PU_3_1_n1_1 | 0.80290495 | 0.802904951323892 | 1.0000e-04 | 5.00000000 | False | m_PU_SG_PU_3_n1_1_1 |
| m_SU_SG_SU_1_0_0_1 | 2.68606212 | 2.686062120382649 | 1.0000e-04 | 5.00000000 | True | |
| m_SU_SG_SU_3_0_0_1 | 1.10915311 | 1.109153108617096 | 1.0000e-04 | 5.00000000 | True | |
| p_PU_SG_PU_1_n1_1_1 | -0.86104140 | -0.8610414024232179 | -3.14159265 | 3.14159265 | False | |
| p_PU_SG_PU_1_1_n1_1 | -0.86104140 | -0.8610414024232179 | -3.14159265 | 3.14159265 | False | p_PU_SG_PU_1_n1_1_1 |
| p_PU_SG_PU_3_n1_1_1 | -3.12044446 | -3.1204444620772467 | -3.14159265 | 3.14159265 | True | |
| p_PU_SG_PU_3_1_n1_1 | -3.12044446 | -3.1204444620772467 | -3.14159265 | 3.14159265 | False | p_PU_SG_PU_3_n1_1_1 |
| p_SU_SG_SU_1_0_0_1 | 2.61122920 | 2.611229196458127 | -3.14159265 | 3.14159265 | True | |
| p_SU_SG_SU_3_0_0_1 | -0.07867828 | -0.07867827542158025 | -3.14159265 | 3.14159265 | True |
[23]:
# Plot params violin style
# data.paramPlot(dataType='m', sel = 'E', hue = 'redchi', backend='hv', hvType='violin', returnFlag = True, remap = 'lmMap')
data.paramPlot(selectors={'Type':'m', 'redchiGroup':'E'}, hue = 'redchi', backend='hv', hvType='violin', returnFlag = True, remap = 'lmMap')
mParams = data.data['plots']['paramPlot']
data.paramPlot(selectors={'Type':'p', 'redchiGroup':'E'}, hue = 'redchi', backend='hv', hvType='violin', returnFlag = True, remap = 'lmMap')
pParams = data.data['plots']['paramPlot']
*** Warning: found MultiIndex for DataFrame data.index - checkDims may have issues with Pandas MultiIndex, but will try anyway.
*** Warning: found MultiIndex for DataFrame data.index - checkDims may have issues with Pandas MultiIndex, but will try anyway.
[24]:
# Replot as 2-up layout
# Setting shared_axes=False works, although also looses link over x-axis.
# Need to rename dims? See https://github.com/holoviz/holoviews/issues/714
# E.g. mParams.redim(y={'name','Mag'}) # Works for label, but not underlying plots
layout3 = (mParams.redim(Mag=hv.Dimension('Mag', range=(1, 3))).opts(frame_width = figSize[0], frame_height = figSize[1], xaxis=None, ylabel='Mag', ylim=(1.0,3.0), title='(a) Magnitudes') +
pParams.opts(frame_width = figSize[0], frame_height = figSize[1], ylabel='Phase', title='(b) Phases')).opts(shared_axes=False).cols(1)
pType = 'paramsViolin'
data.data['plots'][pType] = layout3
if saveFigs:
data.hvSave(pTypes = pType)
# Show plot
layout3
Saving self.data[plots][paramsViolin] to D:\code\python\jupyterNotebooks\dev\PEMtk\fitting\multiFit_tests_and_parallel\figs_290722\dataDump_1000fitTests_multiFit_noise_051021_paramsViolin_290722_11-26-33.png
Saving self.data[plots][paramsViolin] to D:\code\python\jupyterNotebooks\dev\PEMtk\fitting\multiFit_tests_and_parallel\figs_290722\dataDump_1000fitTests_multiFit_noise_051021_paramsViolin_290722_11-26-35.html
[24]:
Rebin over further reduced range - Correlation plot & \(\beta_{LM}\) plots
[25]:
# Set new classifiers for small range
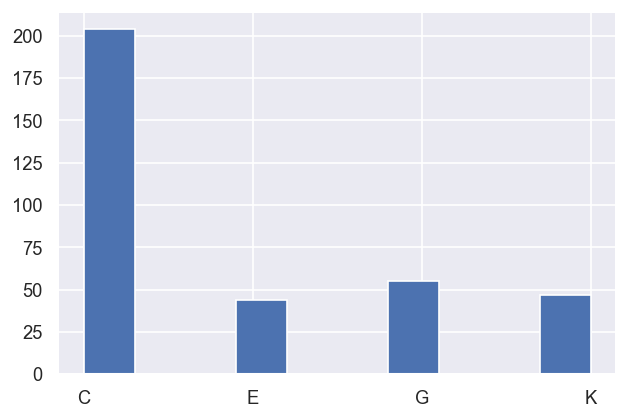
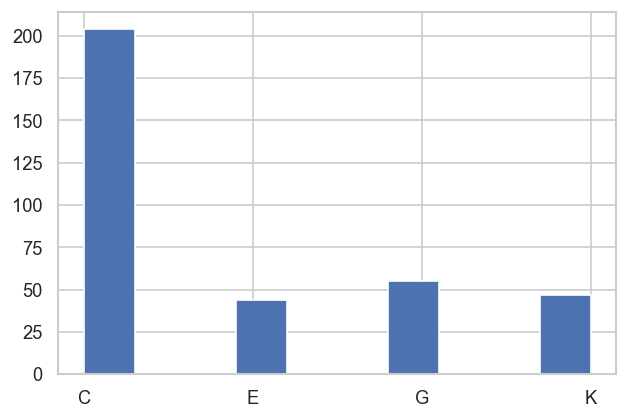
data.classifyFits(bins = [2.29e-4, 2.3e-4,20])
| success | chisqr | redchi | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| count | unique | top | freq | count | unique | top | freq | count | unique | top | freq | |
| redchiGroup | ||||||||||||
| A | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN |
| B | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN |
| C | 204 | 1 | True | 204 | 204.000 | 204.000 | 0.042 | 1.000 | 204.000 | 204.000 | 0.000 | 1.000 |
| D | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN |
| E | 44 | 1 | True | 44 | 44.000 | 44.000 | 0.042 | 1.000 | 44.000 | 44.000 | 0.000 | 1.000 |
| F | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN |
| G | 55 | 1 | True | 55 | 55.000 | 55.000 | 0.042 | 1.000 | 55.000 | 55.000 | 0.000 | 1.000 |
| H | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN |
| I | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN |
| J | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN |
| K | 47 | 1 | True | 47 | 47.000 | 47.000 | 0.042 | 1.000 | 47.000 | 47.000 | 0.000 | 1.000 |
| L | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN |
| M | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN |
| N | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN |
| O | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN |
| P | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN |
| Q | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN |
| R | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN |
| S | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN |
*** Warning: found MultiIndex for DataFrame data.index - checkDims may have issues with Pandas MultiIndex, but will try anyway.
*** Warning: found MultiIndex for DataFrame data.index - checkDims may have issues with Pandas MultiIndex, but will try anyway.
[26]:
# corrPlot() with subselections (two levels)
# NOTE: with hue mapping for a continuous range this can be quite slow (minutes).
# Additional style forcing... not picking up globals here?
hvPlotters.sns.set_style("whitegrid") # OK
# no phase corr
# data.corrPlot(dataType = 'C', level = 'redchiGroup', sel = 'p', selLevel = 'Type', hue = 'redchi')
# With phase corr
data.phaseCorrection(absFlag=True,useRef=False) # NOTE - need to rerun phaseCorrection after classification
# data.phaseCorrection(useRef=False) # Ref to self. (Defaults to input matrix elements)
# data.paramPlot(dataType='pc', sel = 'C', hue = 'redchi', backend='hv', hRound = 9)
# data.corrPlot(dataType = 'C', level = 'redchiGroup', sel = 'pc', selLevel = 'Type', hue = 'redchi') # Check pair correlations including redchi
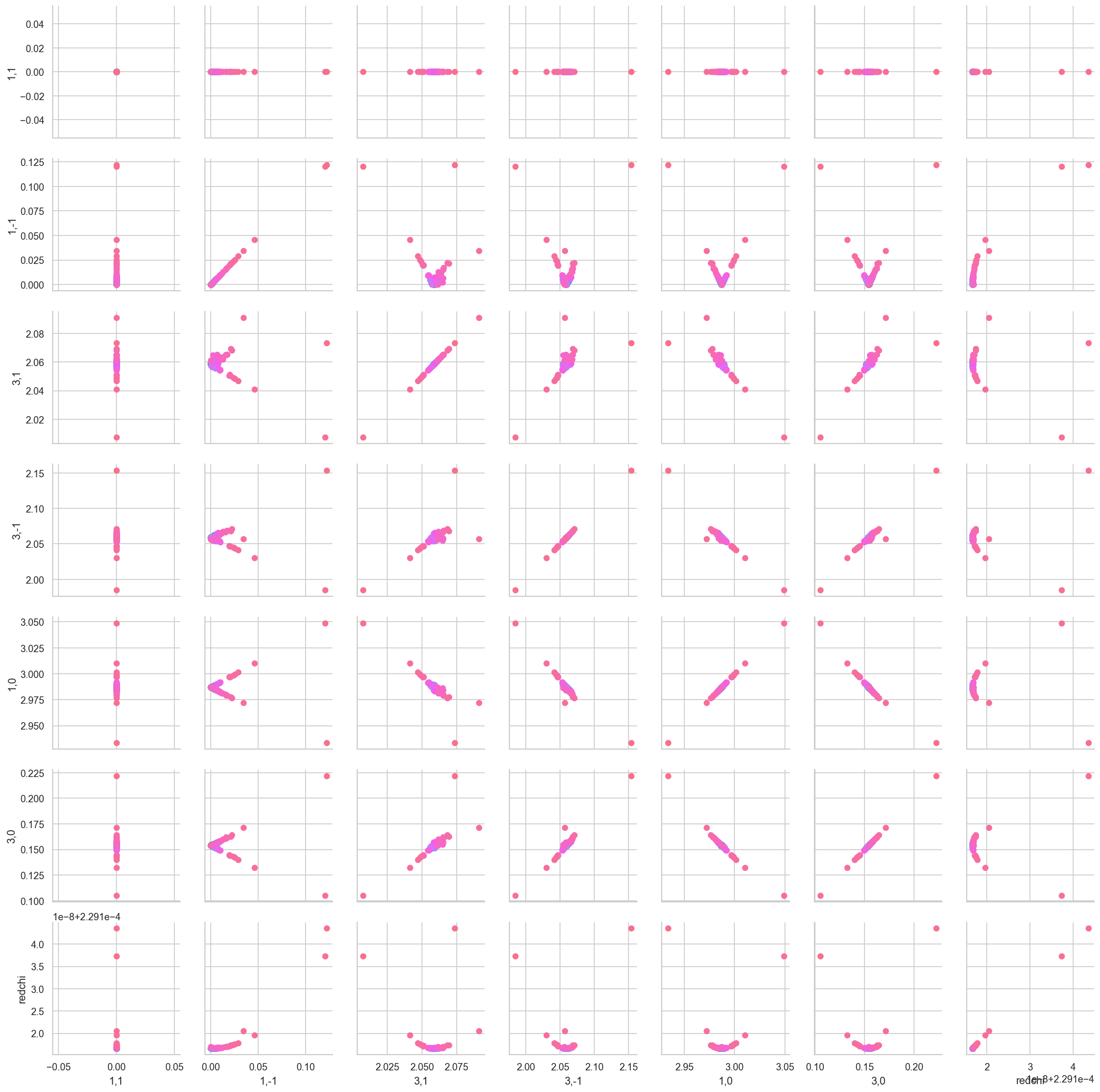
data.corrPlot(dataType = 'C', level = 'redchiGroup', sel = 'pc', selLevel = 'Type', hue = 'redchi', remap = 'lmMap') #, alpha = 0.3)
# plot_kws={'alpha': 0.1}) # Additional args for Seaborn PairGrid, see https://stackoverflow.com/questions/47200033/how-to-adjust-transparency-alpha-in-seaborn-pairplot
# plot_kws={'scatter_kws': {'alpha': 0.1}}) # Additional args
# data.corrPlot(dataType = 'C', level = 'redchiGroup', sel = 'pc', selLevel = 'Type', hue = 'Fit') # Check pair correlations including Fit #
if saveFigs:
# plt.title('')
# plt.ylabel(r'$\beta_{L,M}$')
import matplotlib.pyplot as plt
from datetime import datetime as dt
timeString = dt.now()
plt.savefig(f'{data.job["fileIn"].stem}_corrPlot_{timeString.strftime("%d%m%y")}.png')
*** Warning: found MultiIndex for DataFrame data.index - checkDims may have issues with Pandas MultiIndex, but will try anyway.
*** Warning: found MultiIndex for DataFrame data.index - checkDims may have issues with Pandas MultiIndex, but will try anyway.
Set ref param = PU_SG_PU_1_1_n1_1
| Param | PU_SG_PU_1_1_n1_1 | PU_SG_PU_1_n1_1_1 | PU_SG_PU_3_1_n1_1 | PU_SG_PU_3_n1_1_1 | SU_SG_SU_1_0_0_1 | SU_SG_SU_3_0_0_1 | ||
|---|---|---|---|---|---|---|---|---|
| Fit | Type | redchiGroup | ||||||
| 2 | m | C | 1.610 | 1.618 | 1.141 | 1.144 | 2.653 | 1.144 |
| n | C | 0.400 | 0.402 | 0.284 | 0.284 | 0.660 | 0.285 | |
| p | C | -0.857 | -0.861 | 1.199 | 1.200 | 2.437 | -0.704 | |
| pc | C | 0.000 | 0.004 | 2.056 | 2.057 | 2.989 | 0.153 | |
| 8 | m | G | 1.569 | 1.655 | 1.167 | 1.207 | 2.660 | 1.130 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 997 | pc | C | 0.000 | 0.002 | 2.059 | 2.061 | 2.986 | 0.155 |
| 999 | m | C | 1.610 | 1.617 | 1.141 | 1.144 | 2.653 | 1.144 |
| n | C | 0.400 | 0.402 | 0.284 | 0.285 | 0.660 | 0.285 | |
| p | C | -0.856 | -0.861 | 1.200 | 1.200 | 2.438 | -0.704 | |
| pc | C | 0.000 | 0.005 | 2.056 | 2.056 | 2.990 | 0.152 |
1400 rows × 6 columns
<seaborn.axisgrid.PairGrid at 0x11aba4629b0>
[27]:
# Get parameter statistics.
# Specify subselection criteria with a dictionary of selectors (currently uses index values only)
# TODO: more general selection & groupings here.
data.paramsReport(inds = {'redchiGroup':'C'})
*** Warning: found MultiIndex for DataFrame data.index - checkDims may have issues with Pandas MultiIndex, but will try anyway.
Set parameter stats to self.paramsSummary.
| Type | m | n | p | pc | |
|---|---|---|---|---|---|
| Param | Agg | ||||
| PU_SG_PU_1_1_n1_1 | min | 1.605 | 0.399 | -0.883 | 0.000 |
| mean | 1.614 | 0.401 | -0.859 | 0.000 | |
| median | 1.614 | 0.401 | -0.861 | 0.000 | |
| max | 1.626 | 0.404 | -0.739 | 0.000 | |
| std | 0.002 | 0.001 | 0.014 | 0.000 | |
| var | 0.000 | 0.000 | 0.000 | 0.000 | |
| PU_SG_PU_1_n1_1_1 | min | 1.602 | 0.398 | -0.861 | 0.000 |
| mean | 1.614 | 0.401 | -0.861 | 0.005 | |
| median | 1.614 | 0.401 | -0.861 | 0.002 | |
| max | 1.628 | 0.404 | -0.861 | 0.122 | |
| std | 0.002 | 0.001 | 0.000 | 0.013 | |
| var | 0.000 | 0.000 | 0.000 | 0.000 | |
| PU_SG_PU_3_1_n1_1 | min | 1.124 | 0.279 | -2.931 | 2.007 |
| mean | 1.143 | 0.284 | 0.149 | 2.059 | |
| median | 1.143 | 0.284 | 1.197 | 2.059 | |
| max | 1.174 | 0.292 | 1.266 | 2.091 | |
| std | 0.003 | 0.001 | 1.798 | 0.005 | |
| var | 0.000 | 0.000 | 3.232 | 0.000 | |
| PU_SG_PU_3_n1_1_1 | min | 1.124 | 0.279 | -2.928 | 1.985 |
| mean | 1.142 | 0.284 | 0.149 | 2.059 | |
| median | 1.142 | 0.284 | 1.197 | 2.059 | |
| max | 1.173 | 0.291 | 1.244 | 2.154 | |
| std | 0.003 | 0.001 | 1.798 | 0.009 | |
| var | 0.000 | 0.000 | 3.234 | 0.000 | |
| SU_SG_SU_1_0_0_1 | min | 2.652 | 0.659 | 2.116 | 2.934 |
| mean | 2.653 | 0.660 | 2.357 | 2.987 | |
| median | 2.653 | 0.660 | 2.434 | 2.987 | |
| max | 2.653 | 0.660 | 2.493 | 3.049 | |
| std | 0.000 | 0.000 | 0.134 | 0.007 | |
| var | 0.000 | 0.000 | 0.018 | 0.000 | |
| SU_SG_SU_3_0_0_1 | min | 1.143 | 0.284 | -1.026 | 0.105 |
| mean | 1.144 | 0.285 | -0.785 | 0.154 | |
| median | 1.144 | 0.285 | -0.708 | 0.154 | |
| max | 1.144 | 0.285 | -0.636 | 0.222 | |
| std | 0.000 | 0.000 | 0.134 | 0.007 | |
| var | 0.000 | 0.000 | 0.018 | 0.000 |
[28]:
# The default routine aggreagates over all fits (mean + std for spread) and stacks plots
# Set xDim to ensure consistent plotting
data.BLMsetPlot(xDim = 't')
[29]:
# Replot + input data
figSize = [700,600]
layout = (hvBLMPlot * data.data['plots']['BLMsetPlot']).opts(frame_width = figSize[0], frame_height = figSize[1], xaxis=True)
# hvPlotters.hv.save(layout3, f'{fName.stem}_BLM_allFits_150422', fmt='html')
# hvPlotters.hv.save(layout3, f'{fName.stem}_BLM_allFits_150422', fmt='png')
pType = 'BLMdataAgg'
data.data['plots'][pType] = layout
if saveFigs:
data.hvSave(pTypes = pType)
layout
Saving self.data[plots][BLMdataAgg] to D:\code\python\jupyterNotebooks\dev\PEMtk\fitting\multiFit_tests_and_parallel\figs_290722\dataDump_1000fitTests_multiFit_noise_051021_BLMdataAgg_290722_11-29-50.png
Saving self.data[plots][BLMdataAgg] to D:\code\python\jupyterNotebooks\dev\PEMtk\fitting\multiFit_tests_and_parallel\figs_290722\dataDump_1000fitTests_multiFit_noise_051021_BLMdataAgg_290722_11-29-53.html
[29]:
Tables
Final parameters
[30]:
# Note - may need to rerun classifier here to ensure correct dataset if messed about with above!
data.classifyFits(bins = [2.29e-4, 2.3e-4,20]) # Rerun classifier to clean-up
data.phaseCorrection(absFlag=True, useRef = False)
data.paramsReport(inds = {'redchiGroup':'C'})
| success | chisqr | redchi | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| count | unique | top | freq | count | unique | top | freq | count | unique | top | freq | |
| redchiGroup | ||||||||||||
| A | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN |
| B | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN |
| C | 204 | 1 | True | 204 | 204.000 | 204.000 | 0.042 | 1.000 | 204.000 | 204.000 | 0.000 | 1.000 |
| D | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN |
| E | 44 | 1 | True | 44 | 44.000 | 44.000 | 0.042 | 1.000 | 44.000 | 44.000 | 0.000 | 1.000 |
| F | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN |
| G | 55 | 1 | True | 55 | 55.000 | 55.000 | 0.042 | 1.000 | 55.000 | 55.000 | 0.000 | 1.000 |
| H | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN |
| I | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN |
| J | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN |
| K | 47 | 1 | True | 47 | 47.000 | 47.000 | 0.042 | 1.000 | 47.000 | 47.000 | 0.000 | 1.000 |
| L | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN |
| M | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN |
| N | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN |
| O | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN |
| P | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN |
| Q | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN |
| R | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN |
| S | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN | 0 | 0 | NaN | NaN |
*** Warning: found MultiIndex for DataFrame data.index - checkDims may have issues with Pandas MultiIndex, but will try anyway.
*** Warning: found MultiIndex for DataFrame data.index - checkDims may have issues with Pandas MultiIndex, but will try anyway.
*** Warning: found MultiIndex for DataFrame data.index - checkDims may have issues with Pandas MultiIndex, but will try anyway.
*** Warning: found MultiIndex for DataFrame data.index - checkDims may have issues with Pandas MultiIndex, but will try anyway.
Set ref param = PU_SG_PU_1_1_n1_1
| Param | PU_SG_PU_1_1_n1_1 | PU_SG_PU_1_n1_1_1 | PU_SG_PU_3_1_n1_1 | PU_SG_PU_3_n1_1_1 | SU_SG_SU_1_0_0_1 | SU_SG_SU_3_0_0_1 | ||
|---|---|---|---|---|---|---|---|---|
| Fit | Type | redchiGroup | ||||||
| 2 | m | C | 1.610 | 1.618 | 1.141 | 1.144 | 2.653 | 1.144 |
| n | C | 0.400 | 0.402 | 0.284 | 0.284 | 0.660 | 0.285 | |
| p | C | -0.857 | -0.861 | 1.199 | 1.200 | 2.437 | -0.704 | |
| pc | C | 0.000 | 0.004 | 2.056 | 2.057 | 2.989 | 0.153 | |
| 8 | m | G | 1.569 | 1.655 | 1.167 | 1.207 | 2.660 | 1.130 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 997 | pc | C | 0.000 | 0.002 | 2.059 | 2.061 | 2.986 | 0.155 |
| 999 | m | C | 1.610 | 1.617 | 1.141 | 1.144 | 2.653 | 1.144 |
| n | C | 0.400 | 0.402 | 0.284 | 0.285 | 0.660 | 0.285 | |
| p | C | -0.856 | -0.861 | 1.200 | 1.200 | 2.438 | -0.704 | |
| pc | C | 0.000 | 0.005 | 2.056 | 2.056 | 2.990 | 0.152 |
1400 rows × 6 columns
*** Warning: found MultiIndex for DataFrame data.index - checkDims may have issues with Pandas MultiIndex, but will try anyway.
Set parameter stats to self.paramsSummary.
| Type | m | n | p | pc | |
|---|---|---|---|---|---|
| Param | Agg | ||||
| PU_SG_PU_1_1_n1_1 | min | 1.605 | 0.399 | -0.883 | 0.000 |
| mean | 1.614 | 0.401 | -0.859 | 0.000 | |
| median | 1.614 | 0.401 | -0.861 | 0.000 | |
| max | 1.626 | 0.404 | -0.739 | 0.000 | |
| std | 0.002 | 0.001 | 0.014 | 0.000 | |
| var | 0.000 | 0.000 | 0.000 | 0.000 | |
| PU_SG_PU_1_n1_1_1 | min | 1.602 | 0.398 | -0.861 | 0.000 |
| mean | 1.614 | 0.401 | -0.861 | 0.005 | |
| median | 1.614 | 0.401 | -0.861 | 0.002 | |
| max | 1.628 | 0.404 | -0.861 | 0.122 | |
| std | 0.002 | 0.001 | 0.000 | 0.013 | |
| var | 0.000 | 0.000 | 0.000 | 0.000 | |
| PU_SG_PU_3_1_n1_1 | min | 1.124 | 0.279 | -2.931 | 2.007 |
| mean | 1.143 | 0.284 | 0.149 | 2.059 | |
| median | 1.143 | 0.284 | 1.197 | 2.059 | |
| max | 1.174 | 0.292 | 1.266 | 2.091 | |
| std | 0.003 | 0.001 | 1.798 | 0.005 | |
| var | 0.000 | 0.000 | 3.232 | 0.000 | |
| PU_SG_PU_3_n1_1_1 | min | 1.124 | 0.279 | -2.928 | 1.985 |
| mean | 1.142 | 0.284 | 0.149 | 2.059 | |
| median | 1.142 | 0.284 | 1.197 | 2.059 | |
| max | 1.173 | 0.291 | 1.244 | 2.154 | |
| std | 0.003 | 0.001 | 1.798 | 0.009 | |
| var | 0.000 | 0.000 | 3.234 | 0.000 | |
| SU_SG_SU_1_0_0_1 | min | 2.652 | 0.659 | 2.116 | 2.934 |
| mean | 2.653 | 0.660 | 2.357 | 2.987 | |
| median | 2.653 | 0.660 | 2.434 | 2.987 | |
| max | 2.653 | 0.660 | 2.493 | 3.049 | |
| std | 0.000 | 0.000 | 0.134 | 0.007 | |
| var | 0.000 | 0.000 | 0.018 | 0.000 | |
| SU_SG_SU_3_0_0_1 | min | 1.143 | 0.284 | -1.026 | 0.105 |
| mean | 1.144 | 0.285 | -0.785 | 0.154 | |
| median | 1.144 | 0.285 | -0.708 | 0.154 | |
| max | 1.144 | 0.285 | -0.636 | 0.222 | |
| std | 0.000 | 0.000 | 0.134 | 0.007 | |
| var | 0.000 | 0.000 | 0.018 | 0.000 |
[31]:
# With abs phaseCorrection flag set
import pandas as pd
# pd.options.display.precision = 5
pd.options.display.float_format = '{:.3f}'.format
data.paramsCompare(phaseCorrParams={'absFlag':True, 'useRef':False})
*** Warning: found MultiIndex for DataFrame data.index - checkDims may have issues with Pandas MultiIndex, but will try anyway.
*** Warning: found MultiIndex for DataFrame data.index - checkDims may have issues with Pandas MultiIndex, but will try anyway.
Set ref param = PU_SG_PU_1_1_n1_1
| Param | PU_SG_PU_1_1_n1_1 | PU_SG_PU_1_n1_1_1 | PU_SG_PU_3_1_n1_1 | PU_SG_PU_3_n1_1_1 | SU_SG_SU_1_0_0_1 | SU_SG_SU_3_0_0_1 | |
|---|---|---|---|---|---|---|---|
| Fit | Type | ||||||
| ref | m | 1.785 | 1.785 | 0.803 | 0.803 | 2.686 | 1.109 |
| n | 0.445 | 0.445 | 0.200 | 0.200 | 0.669 | 0.276 | |
| p | -0.861 | -0.861 | -3.120 | -3.120 | 2.611 | -0.079 | |
| pc | 0.000 | 0.000 | 2.259 | 2.259 | 2.811 | 0.782 |
Set parameter comparison to self.paramsSummaryComp.
| Param | PU_SG_PU_1_1_n1_1 | PU_SG_PU_1_n1_1_1 | PU_SG_PU_3_1_n1_1 | PU_SG_PU_3_n1_1_1 | SU_SG_SU_1_0_0_1 | SU_SG_SU_3_0_0_1 | ||
|---|---|---|---|---|---|---|---|---|
| Type | Source | dType | ||||||
| m | mean | num | 1.614 | 1.614 | 1.143 | 1.142 | 2.653 | 1.144 |
| ref | num | 1.785 | 1.785 | 0.803 | 0.803 | 2.686 | 1.109 | |
| diff | % | 10.583 | 10.599 | 29.733 | 29.715 | 1.264 | 3.030 | |
| num | -0.171 | -0.171 | 0.340 | 0.339 | -0.034 | 0.035 | ||
| std | % | 0.130 | 0.137 | 0.266 | 0.265 | 0.002 | 0.007 | |
| num | 0.002 | 0.002 | 0.003 | 0.003 | 0.000 | 0.000 | ||
| diff/std | % | 8139.354 | 7726.095 | 11168.316 | 11210.075 | 79267.770 | 44813.500 | |
| n | mean | num | 0.401 | 0.401 | 0.284 | 0.284 | 0.660 | 0.285 |
| ref | num | 0.445 | 0.445 | 0.200 | 0.200 | 0.669 | 0.276 | |
| diff | % | 10.784 | 10.801 | 29.604 | 29.587 | 1.449 | 2.853 | |
| num | -0.043 | -0.043 | 0.084 | 0.084 | -0.010 | 0.008 | ||
| std | % | 0.133 | 0.133 | 0.262 | 0.264 | 0.014 | 0.018 | |
| num | 0.001 | 0.001 | 0.001 | 0.001 | 0.000 | 0.000 | ||
| diff/std | % | 8123.972 | 8123.937 | 11298.199 | 11228.378 | 10459.719 | 15923.757 | |
| p | mean | num | -0.859 | -0.861 | 0.149 | 0.149 | 2.357 | -0.785 |
| ref | num | -0.861 | -0.861 | -3.120 | -3.120 | 2.611 | -0.079 | |
| diff | % | 0.195 | 0.000 | 2189.833 | 2198.785 | 10.779 | 89.974 | |
| num | 0.002 | 0.000 | 3.270 | 3.269 | -0.254 | -0.706 | ||
| std | % | 1.632 | 0.000 | 1204.022 | 1209.452 | 5.699 | 17.083 | |
| num | 0.014 | 0.000 | 1.798 | 1.798 | 0.134 | 0.134 | ||
| diff/std | % | 11.953 | inf | 181.876 | 181.800 | 189.129 | 526.696 | |
| pc | mean | num | 0.000 | 0.005 | 2.059 | 2.059 | 2.987 | 0.154 |
| ref | num | 0.000 | 0.000 | 2.259 | 2.259 | 2.811 | 0.782 | |
| diff | % | nan | 100.000 | 9.741 | 9.738 | 5.893 | 407.161 | |
| num | 0.000 | 0.005 | -0.201 | -0.200 | 0.176 | -0.628 | ||
| std | % | nan | 243.149 | 0.256 | 0.460 | 0.226 | 4.424 | |
| num | 0.000 | 0.013 | 0.005 | 0.009 | 0.007 | 0.007 | ||
| diff/std | % | nan | 41.127 | 3798.755 | 2114.926 | 2605.648 | 9204.066 |
[32]:
# Output table to latex
if saveTables:
data.paramsSummaryComp.to_latex(f'paramsComp_latex_{timeString.strftime("%d%m%y")}.tex')
Init & final relabelled
Aggregate results from best fits
[33]:
data.setAggMatE(simpleForm = True)
Set reformatted aggregate data to self.data[agg][matEpd].
[34]:
data.data['agg']['matEpd']
[34]:
| Type | m | n | p | pc | comp | compC | labels | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Cont | l | m | mu | |||||||
| PU | 1 | -1 | 1 | 1.614 | 0.401 | -0.859 | 0.000 | 1.054-1.222j | 0.401+0.000j | 1,-1 |
| 1 | -1 | 1.614 | 0.401 | -0.861 | 0.005 | 1.051-1.224j | 0.401+0.002j | 1,1 | ||
| 3 | -1 | 1 | 1.143 | 0.284 | 0.149 | 2.059 | 1.130+0.170j | -0.133+0.251j | 3,-1 | |
| 1 | -1 | 1.142 | 0.284 | 0.149 | 2.059 | 1.130+0.169j | -0.133+0.251j | 3,1 | ||
| SU | 1 | 0 | 0 | 2.653 | 0.660 | 2.357 | 2.987 | -1.877+1.874j | -0.652+0.102j | 1,0 |
| 3 | 0 | 0 | 1.144 | 0.285 | -0.785 | 0.154 | 0.809-0.808j | 0.281+0.044j | 3,0 |
[35]:
if saveTables:
data.data['agg']['matEpd'].to_latex(f'paramsAggRelabelled_latex_{timeString.strftime("%d%m%y")}.tex')
Input matrix elements
Use setAggMatE for consistent reformatting to table.
[36]:
data.setAggMatE(key = 'ref', simpleForm = True)
data.data['ref']['matEpd']
Set reformatted ref data to self.data[ref][matEpd].
[36]:
| comp | m | p | labels | ||||
|---|---|---|---|---|---|---|---|
| Cont | l | m | mu | ||||
| PU | 1 | -1 | 1 | 1.163-1.354j | 1.785 | -0.861 | 1,-1 |
| 1 | -1 | 1.163-1.354j | 1.785 | -0.861 | 1,1 | ||
| 3 | -1 | 1 | -0.803-0.017j | 0.803 | -3.120 | 3,-1 | |
| 1 | -1 | -0.803-0.017j | 0.803 | -3.120 | 3,1 | ||
| SU | 1 | 0 | 0 | -2.317+1.359j | 2.686 | 2.611 | 1,0 |
| 3 | 0 | 0 | 1.106-0.087j | 1.109 | -0.079 | 3,0 |
[37]:
if saveTables:
data.data['agg']['matEpd'].to_latex(f'paramsInputRelabelled_latex_{timeString.strftime("%d%m%y")}.tex')
Comparison table relabelled
Update table with renamed params from list.
[38]:
summaryRenamed = pemtk.fit._util.renameParams(data.paramsSummaryComp, data.lmmu['lmMap'])
summaryRenamed
[38]:
| lm | 1,1 | 1,-1 | 3,1 | 3,-1 | 1,0 | 3,0 | ||
|---|---|---|---|---|---|---|---|---|
| Type | Source | dType | ||||||
| m | mean | num | 1.614 | 1.614 | 1.143 | 1.142 | 2.653 | 1.144 |
| ref | num | 1.785 | 1.785 | 0.803 | 0.803 | 2.686 | 1.109 | |
| diff | % | 10.583 | 10.599 | 29.733 | 29.715 | 1.264 | 3.030 | |
| num | -0.171 | -0.171 | 0.340 | 0.339 | -0.034 | 0.035 | ||
| std | % | 0.130 | 0.137 | 0.266 | 0.265 | 0.002 | 0.007 | |
| num | 0.002 | 0.002 | 0.003 | 0.003 | 0.000 | 0.000 | ||
| diff/std | % | 8139.354 | 7726.095 | 11168.316 | 11210.075 | 79267.770 | 44813.500 | |
| n | mean | num | 0.401 | 0.401 | 0.284 | 0.284 | 0.660 | 0.285 |
| ref | num | 0.445 | 0.445 | 0.200 | 0.200 | 0.669 | 0.276 | |
| diff | % | 10.784 | 10.801 | 29.604 | 29.587 | 1.449 | 2.853 | |
| num | -0.043 | -0.043 | 0.084 | 0.084 | -0.010 | 0.008 | ||
| std | % | 0.133 | 0.133 | 0.262 | 0.264 | 0.014 | 0.018 | |
| num | 0.001 | 0.001 | 0.001 | 0.001 | 0.000 | 0.000 | ||
| diff/std | % | 8123.972 | 8123.937 | 11298.199 | 11228.378 | 10459.719 | 15923.757 | |
| p | mean | num | -0.859 | -0.861 | 0.149 | 0.149 | 2.357 | -0.785 |
| ref | num | -0.861 | -0.861 | -3.120 | -3.120 | 2.611 | -0.079 | |
| diff | % | 0.195 | 0.000 | 2189.833 | 2198.785 | 10.779 | 89.974 | |
| num | 0.002 | 0.000 | 3.270 | 3.269 | -0.254 | -0.706 | ||
| std | % | 1.632 | 0.000 | 1204.022 | 1209.452 | 5.699 | 17.083 | |
| num | 0.014 | 0.000 | 1.798 | 1.798 | 0.134 | 0.134 | ||
| diff/std | % | 11.953 | inf | 181.876 | 181.800 | 189.129 | 526.696 | |
| pc | mean | num | 0.000 | 0.005 | 2.059 | 2.059 | 2.987 | 0.154 |
| ref | num | 0.000 | 0.000 | 2.259 | 2.259 | 2.811 | 0.782 | |
| diff | % | nan | 100.000 | 9.741 | 9.738 | 5.893 | 407.161 | |
| num | 0.000 | 0.005 | -0.201 | -0.200 | 0.176 | -0.628 | ||
| std | % | nan | 243.149 | 0.256 | 0.460 | 0.226 | 4.424 | |
| num | 0.000 | 0.013 | 0.005 | 0.009 | 0.007 | 0.007 | ||
| diff/std | % | nan | 41.127 | 3798.755 | 2114.926 | 2605.648 | 9204.066 |
[39]:
if saveTables:
data.data['agg']['matEpd'].to_latex(f'paramsInputRelabelled_latex_{timeString.strftime("%d%m%y")}.tex')
MFPAD plotting
FINAL PLOTTING IN MFPAD_replotting_from_file_190722-dist.ipynb for tidy version
UPDATES here for data handling and export only.
[40]:
# Set Agg data to XR dataset, along with ref data set
data.aggToXR(refKey = 'orb5', returnType = 'ds', conformDims=True) # OK
# data.aggToXR(refKey = 'orb5', returnType = 'ds', conformDims=False) # If False, set to dict, otherwise fails at DS stacking
# data.aggToXR(refKey = 'orb5', returnType = 'da', conformDims=False) # OK - sets to separate DAs. May want this for better dim handling - missing some sym dims in current example case.
# data.aggToXR(returnType = 'ds', conformDims=False) # OK without refKey to stack with different dims.
Added dim Targ
Added dim Total
Added dim Targ
Added dim Total
Set XR dataset for self.data['agg']['matE']
[41]:
data.data['agg']['matE']
[41]:
- Eke: 51
- LM: 18
- Sym: 4
- Type: 4
- it: 1
- mu: 3
- Type(Type)object'L' 'V' 'comp' 'compC'
array(['L', 'V', 'comp', 'compC'], dtype=object)
- LM(LM)MultiIndex(l, m)
array([(1, -1), (1, 0), (1, 1), (3, -1), (3, 0), (3, 1), (5, -1), (5, 0), (5, 1), (7, -1), (7, 0), (7, 1), (9, -1), (9, 0), (9, 1), (11, -1), (11, 0), (11, 1)], dtype=object) - l(LM)int641 1 1 3 3 3 5 ... 7 9 9 9 11 11 11
array([ 1, 1, 1, 3, 3, 3, 5, 5, 5, 7, 7, 7, 9, 9, 9, 11, 11, 11], dtype=int64) - m(LM)int64-1 0 1 -1 0 1 -1 ... -1 0 1 -1 0 1
array([-1, 0, 1, -1, 0, 1, -1, 0, 1, -1, 0, 1, -1, 0, 1, -1, 0, 1], dtype=int64) - Sym(Sym)MultiIndex(Cont, Targ, Total)
array([('PU', 'SG', 'PU'), ('PU', 'U', 'U'), ('SU', 'SG', 'SU'), ('SU', 'U', 'U')], dtype=object) - Cont(Sym)object'PU' 'PU' 'SU' 'SU'
array(['PU', 'PU', 'SU', 'SU'], dtype=object)
- Targ(Sym)object'SG' 'U' 'SG' 'U'
array(['SG', 'U', 'SG', 'U'], dtype=object)
- Total(Sym)object'PU' 'U' 'SU' 'U'
array(['PU', 'U', 'SU', 'U'], dtype=object)
- mu(mu)int64-1 0 1
array([-1, 0, 1], dtype=int64)
- Ehv(Eke)float6417.4 18.4 19.4 ... 65.4 66.4 67.4
array([17.4, 18.4, 19.4, 20.4, 21.4, 22.4, 23.4, 24.4, 25.4, 26.4, 27.4, 28.4, 29.4, 30.4, 31.4, 32.4, 33.4, 34.4, 35.4, 36.4, 37.4, 38.4, 39.4, 40.4, 41.4, 42.4, 43.4, 44.4, 45.4, 46.4, 47.4, 48.4, 49.4, 50.4, 51.4, 52.4, 53.4, 54.4, 55.4, 56.4, 57.4, 58.4, 59.4, 60.4, 61.4, 62.4, 63.4, 64.4, 65.4, 66.4, 67.4]) - Eke(Eke)float640.1 1.1 2.1 3.1 ... 48.1 49.1 50.1
array([ 0.1, 1.1, 2.1, 3.1, 4.1, 5.1, 6.1, 7.1, 8.1, 9.1, 10.1, 11.1, 12.1, 13.1, 14.1, 15.1, 16.1, 17.1, 18.1, 19.1, 20.1, 21.1, 22.1, 23.1, 24.1, 25.1, 26.1, 27.1, 28.1, 29.1, 30.1, 31.1, 32.1, 33.1, 34.1, 35.1, 36.1, 37.1, 38.1, 39.1, 40.1, 41.1, 42.1, 43.1, 44.1, 45.1, 46.1, 47.1, 48.1, 49.1, 50.1]) - it(it)int641
array([1], dtype=int64)
- SF(Eke)complex128(2.1560627+3.741674j) ... (4.4127053+1.8281945j)
array([2.1560627+3.741674j , 2.2237521+3.6277801j, 2.2894411+3.5236912j, 2.3532973+3.4280768j, 2.4154658+3.3398459j, 2.476074 +3.2580947j, 2.5352337+3.1820671j, 2.5930439+3.1111249j, 2.6495932+3.0447254j, 2.7049605+2.9824035j, 2.759217 +2.9237583j, 2.812427 +2.8684419j, 2.8646489+2.816151j , 2.9159356+2.7666193j, 2.9663357+2.7196125j, 3.0158937+2.6749231j, 3.0646504+2.6323667j, 3.1126435+2.5917789j, 3.1599077+2.5530124j, 3.2064753+2.5159351j, 3.2523762+2.4804276j, 3.2976383+2.4463822j, 3.3422875+2.4137013j, 3.386348 +2.3822961j, 3.4298425+2.3520857j, 3.4727924+2.3229962j, 3.5152175+2.29496j , 3.5571367+2.267915j , 3.5985676+2.2418041j, 3.6395269+2.2165748j, 3.6800303+2.1921786j, 3.7200928+2.1685705j, 3.7597284+2.1457091j, 3.7989505+2.1235559j, 3.8377718+2.1020749j, 3.8762043+2.0812328j, 3.9142594+2.0609987j, 3.9519481+2.0413435j, 3.9892808+2.0222401j, 4.0262673+2.0036632j, 4.0629171+1.985589j , 4.0992393+1.9679953j, 4.1352425+1.9508611j, 4.1709348+1.9341668j, 4.2063244+1.9178938j, 4.2414186+1.9020249j, 4.2762249+1.8865434j, 4.3107501+1.8714338j, 4.345001 +1.8566817j, 4.378984 +1.8422729j, 4.4127053+1.8281945j])
- comp(Type, mu, LM, Sym)complex128(nan+nanj) ... (nan+nanj)
array([[[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, 1.05149172-1.22394193j, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, 1.12975546+0.16921918j, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, -1.87741993+1.87381355j], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, 0.80933551-0.80825182j], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, 1.05370029-1.22236047j, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, 1.12992788+0.16998089j, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]]]) - compC(Type, mu, LM, Sym)complex128(nan+nanj) ... (nan+nanj)
array([[[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, 0.40135294+0.00215583j, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, -0.13325424+0.25096315j, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, -0.65190516+0.10163353j], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, 0.28112833+0.04371509j], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, 0.40141894+0.j , nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, -0.13327317+0.25103294j, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj], [ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]]]) - orb5(LM, Eke, Sym, mu, it, Type)complex128(nan+nanj) ... (nan+nanj)
- E :
- 0.1
- Ehv :
- 15.68
- SF :
- (2.1560627+3.741674j)
- Lmax :
- 11
- Cont :
- SU
- Targ :
- SG
- Total :
- SU
- QNs :
- ['m', 'l', 'mu', 'ip', 'it', 'Value']
- dataType :
- matE
- file :
- n2_3sg_0.1-50.1eV_A2.inp.out
- fileBase :
- D:\code\github\ePSproc\data\photoionization\n2_multiorb
- fileList :
- n2_3sg_0.1-50.1eV_A2.inp.out
- jobLabel :
- 3sg-1
array([[[[[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[-1.7757076e+00+6.3474768e-01j, -1.9403462e+00+6.9465999e-01j, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]]], [[[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ 1.1629411e+00-1.3536696e+00j, 1.2585400e+00-1.4670488e+00j, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]]], [[[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[-6.7206736e-01-1.5510867e+00j, -7.2345845e-01-1.6647964e+00j, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]]], ..., [[[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[-4.3564182e-01-7.1276154e-01j, -4.2509554e-01-6.6006094e-01j, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]]], [[[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[-4.3153069e-01-7.0462323e-01j, -4.2061423e-01-6.5253773e-01j, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]]], [[[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[-4.2731653e-01-6.9626607e-01j, -4.1606509e-01-6.4490113e-01j, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]]]], [[[[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ 2.7363206e+00-9.2696864e-02j, 2.9815816e+00-1.0062913e-01j, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]]], [[[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[-2.3170597e+00+1.3587362e+00j, -2.4961077e+00+1.4631580e+00j, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]]], [[[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ 1.6888467e-01+2.6389559e+00j, 1.7928455e-01+2.8082277e+00j, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]]], ..., [[[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[-1.3280743e-01+4.3357421e-01j, -1.1225816e-01+3.9916386e-01j, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]]], [[[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[-1.4907960e-01+4.3031896e-01j, -1.2699346e-01+3.9484078e-01j, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]]], [[[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[-1.6461802e-01+4.2668132e-01j, -1.4097903e-01+3.9029174e-01j, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]]]], [[[[[-1.7757076e+00+6.3474768e-01j, -1.9403462e+00+6.9465999e-01j, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]]], [[[[ 1.1629411e+00-1.3536696e+00j, 1.2585400e+00-1.4670488e+00j, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]]], [[[[-6.7206736e-01-1.5510867e+00j, -7.2345845e-01-1.6647964e+00j, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]]], ..., [[[[-4.3564182e-01-7.1276154e-01j, -4.2509554e-01-6.6006094e-01j, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]]], [[[[-4.3153069e-01-7.0462323e-01j, -4.2061423e-01-6.5253773e-01j, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]]], [[[[-4.2731653e-01-6.9626607e-01j, -4.1606509e-01-6.4490113e-01j, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]]]], ..., [[[[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]]], [[[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[-2.2166421e-08+1.4897224e-08j, -2.2462645e-08+1.5296980e-08j, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]]], [[[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ 1.4168322e-07-1.0108773e-07j, 1.4358297e-07-1.0257807e-07j, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]]], ..., [[[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ 9.1652969e-06-3.8486844e-06j, 5.8206861e-06-1.9595570e-06j, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]]], [[[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ 9.0395586e-06-4.2857498e-06j, 5.5904864e-06-2.2352132e-06j, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]]], [[[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ 8.9213389e-06-4.6971505e-06j, 5.3667894e-06-2.4842580e-06j, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]]]], [[[[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]]], [[[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ 3.4737547e-08-1.5180966e-08j, 3.7068537e-08-1.6190679e-08j, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]]], [[[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[-2.2552182e-07+1.1579025e-07j, -2.3947258e-07+1.2304319e-07j, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]]], ..., [[[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[-5.3579641e-06+1.5190377e-06j, -1.5411234e-06-3.5035308e-07j, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]]], [[[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[-5.7787135e-06+1.7595393e-06j, -1.7745738e-06-2.9031071e-07j, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]]], [[[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[-6.1975055e-06+2.0338273e-06j, -2.0070606e-06-2.0324783e-07j, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]]]], [[[[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]]], [[[[-2.2166421e-08+1.4897224e-08j, -2.2462645e-08+1.5296980e-08j, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]]], [[[[ 1.4168322e-07-1.0108773e-07j, 1.4358297e-07-1.0257807e-07j, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]]], ..., [[[[ 9.1652969e-06-3.8486844e-06j, 5.8206861e-06-1.9595570e-06j, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]]], [[[[ 9.0395586e-06-4.2857498e-06j, 5.5904864e-06-2.2352132e-06j, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]]], [[[[ 8.9213389e-06-4.6971505e-06j, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]], [[[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]], [[ nan +nanj, nan +nanj, nan +nanj, nan +nanj]]]]]])
[42]:
# Write data - quick way from dataset
from epsproc.util.listFuncs import dataTypesList
dType = 'matE'
refDims = dataTypesList()[dType]['def']
if exportData:
dataSave = data.data['agg']['matE'].unstack(refDims(sType='sDict'))
dataSave.to_netcdf('matE_fittingAgg_Ref_200722_DS_c.ncf', engine='h5netcdf', invalid_netcdf=True) # OK IF READ IN WITH SAME ENGINE ONLY!
[43]:
# Write data with function + wrapper - TODO
Density matrix plotting
Adapted from https://epsproc.readthedocs.io/en/dev/methods/density_mat_notes_demo_300821.html
[44]:
import numpy as np
def unsignedPhase(da):
"""Convert to unsigned phases."""
# Set mag, phase
mag = da.pipe(np.abs)
phase = da.pipe(np.angle) # Returns np array only!
# Set unsigned
magUS = mag.pipe(np.abs)
# phaseUS = phase.pipe(np.abs)
phaseUS = np.abs(phase)
# Set complex
compFixed = magUS * np.exp(1j* phaseUS)
# return mag,phase
return compFixed
[45]:
# v2 - as v1, but differences for unsigned phase case & fix labels
# 26/07/22: messy but working. Some labelling tricks to push back into matPlot() routine
# Import routines
from epsproc.calc import density
# Compose density matrix
# Set dimensions/state vector/representation
# These must be in original data, but will be restacked as necessary to define the effective basis space.
denDims = ['LM', 'mu']
selDims = {'Type':'L'}
pTypes=['r','i']
thres = 1e-2 # 0.2 # Threshold out l>3 terms if using full 'orb5' set.
normME = False
normDen = 'max'
usPhase = True # Use unsigned phases?
# Calculate - Ref case
# matE = data.data['subset']['matE']
# Set data from master class
# k = 'orb5' # N2 orb5 (SG) dataset
k = 'subset'
matE = data.data[k]['matE']
if normME:
matE = matE/matE.max()
if usPhase:
matE = unsignedPhase(matE)
daOut, *_ = density.densityCalc(matE, denDims = denDims, selDims = selDims, thres = thres) # OK
if normDen=='max':
daOut = daOut/daOut.max()
elif normDen=='trace':
daOut = daOut/(daOut.sum('Sym').pipe(np.trace)**2) # Need sym sum here to get 2D trace
# daPlot = density.matPlot(daOut.sum('Sym'))
daPlot = density.matPlot(daOut.sum('Sym'), pTypes=pTypes)
# Retrieved
matE = data.data['agg']['matE']['compC']
selDims = {'Type':'compC'} # For stacked DS case need to set selDims again here to avoid null data selection below.
if normME:
matE = matE/matE.max()
if usPhase:
matE = unsignedPhase(matE)
daOut2, *_ = density.densityCalc(matE, denDims = denDims, selDims = selDims, thres = thres) # OK
if normDen=='max':
daOut2 = daOut2/daOut2.max()
elif normDen=='trace':
daOut2 = daOut2/(daOut2.sum('Sym').pipe(np.trace)**2)
daPlot2 = density.matPlot(daOut2.sum('Sym'), pTypes=pTypes) #.sel(Eke=slice(0.5,1.5,1)))
# Compute difference
if usPhase:
daDiff = unsignedPhase(daOut.sum('Sym')) - unsignedPhase(daOut2.sum('Sym'))
else:
daDiff = daOut.sum('Sym') - daOut2.sum('Sym')
daDiff.name = 'Difference'
daPlotDiff = density.matPlot(daDiff, pTypes=pTypes)
#******** Plot
daLayout = (daPlot.redim(pType='Component').layout('Component').relabel('(a) Reference density matrix (unsigned phases)') + daPlot2.opts(show_title=False).layout('pType').opts(show_title=True).relabel('(b) Reconstructed') +
daPlotDiff.opts(show_title=False).layout('pType').opts(show_title=True).relabel('(c) Difference')).cols(1)
daLayout.opts(hvPlotters.opts.HeatMap(width=300, frame_width=300, aspect='square', tools=['hover'], colorbar=True, cmap='coolwarm')) # .opts(show_title=False) # .opts(title="Custom Title") #OK
# Notes on titles... see https://holoviews.org/user_guide/Customizing_Plots.html
#
# .relabel('Test') and .opts(title="Custom Title") OK for whole row titles
#
# daPlot2.opts(show_title=False).layout('pType').opts(show_title=True).relabel('Recon') Turns off titles per plot, then titles layout
#
# .redim() to modify individual plot group label (from dimension name)
Set plot kdims to ['l,m,mu', 'l,m,mu_p']; pass kdims = [dim1,dim2] for more control.
Set plot kdims to ['l,m,mu', 'l,m,mu_p']; pass kdims = [dim1,dim2] for more control.
Set plot kdims to ['l,m,mu', 'l,m,mu_p']; pass kdims = [dim1,dim2] for more control.
WARNING:param.HeatMapPlot58525: Due to internal constraints, when aspect and width/height is set, the bokeh backend uses those values as frame_width/frame_height instead. This ensures the aspect is respected, but means that the plot might be slightly larger than anticipated. Set the frame_width/frame_height explicitly to suppress this warning.
WARNING:param.HeatMapPlot58560: Due to internal constraints, when aspect and width/height is set, the bokeh backend uses those values as frame_width/frame_height instead. This ensures the aspect is respected, but means that the plot might be slightly larger than anticipated. Set the frame_width/frame_height explicitly to suppress this warning.
WARNING:param.HeatMapPlot58662: Due to internal constraints, when aspect and width/height is set, the bokeh backend uses those values as frame_width/frame_height instead. This ensures the aspect is respected, but means that the plot might be slightly larger than anticipated. Set the frame_width/frame_height explicitly to suppress this warning.
WARNING:param.HeatMapPlot58697: Due to internal constraints, when aspect and width/height is set, the bokeh backend uses those values as frame_width/frame_height instead. This ensures the aspect is respected, but means that the plot might be slightly larger than anticipated. Set the frame_width/frame_height explicitly to suppress this warning.
WARNING:param.HeatMapPlot58798: Due to internal constraints, when aspect and width/height is set, the bokeh backend uses those values as frame_width/frame_height instead. This ensures the aspect is respected, but means that the plot might be slightly larger than anticipated. Set the frame_width/frame_height explicitly to suppress this warning.
WARNING:param.HeatMapPlot58833: Due to internal constraints, when aspect and width/height is set, the bokeh backend uses those values as frame_width/frame_height instead. This ensures the aspect is respected, but means that the plot might be slightly larger than anticipated. Set the frame_width/frame_height explicitly to suppress this warning.
[45]:
[46]:
# Save plots
pType = f'densityNorm_{normDen}'
data.data['plots'][pType] = daLayout
saveFigs = True
if saveFigs:
data.hvSave(pTypes = pType)
Saving self.data[plots][densityNorm_max] to D:\code\python\jupyterNotebooks\dev\PEMtk\fitting\multiFit_tests_and_parallel\figs_290722\dataDump_1000fitTests_multiFit_noise_051021_densityNorm_max_290722_11-30-03.png
WARNING:param.HeatMapPlot59896: Due to internal constraints, when aspect and width/height is set, the bokeh backend uses those values as frame_width/frame_height instead. This ensures the aspect is respected, but means that the plot might be slightly larger than anticipated. Set the frame_width/frame_height explicitly to suppress this warning.
WARNING:param.HeatMapPlot59931: Due to internal constraints, when aspect and width/height is set, the bokeh backend uses those values as frame_width/frame_height instead. This ensures the aspect is respected, but means that the plot might be slightly larger than anticipated. Set the frame_width/frame_height explicitly to suppress this warning.
WARNING:param.HeatMapPlot60033: Due to internal constraints, when aspect and width/height is set, the bokeh backend uses those values as frame_width/frame_height instead. This ensures the aspect is respected, but means that the plot might be slightly larger than anticipated. Set the frame_width/frame_height explicitly to suppress this warning.
WARNING:param.HeatMapPlot60068: Due to internal constraints, when aspect and width/height is set, the bokeh backend uses those values as frame_width/frame_height instead. This ensures the aspect is respected, but means that the plot might be slightly larger than anticipated. Set the frame_width/frame_height explicitly to suppress this warning.
WARNING:param.HeatMapPlot60169: Due to internal constraints, when aspect and width/height is set, the bokeh backend uses those values as frame_width/frame_height instead. This ensures the aspect is respected, but means that the plot might be slightly larger than anticipated. Set the frame_width/frame_height explicitly to suppress this warning.
WARNING:param.HeatMapPlot60204: Due to internal constraints, when aspect and width/height is set, the bokeh backend uses those values as frame_width/frame_height instead. This ensures the aspect is respected, but means that the plot might be slightly larger than anticipated. Set the frame_width/frame_height explicitly to suppress this warning.
Saving self.data[plots][densityNorm_max] to D:\code\python\jupyterNotebooks\dev\PEMtk\fitting\multiFit_tests_and_parallel\figs_290722\dataDump_1000fitTests_multiFit_noise_051021_densityNorm_max_290722_11-30-09.html
WARNING:param.HeatMapPlot60993: Due to internal constraints, when aspect and width/height is set, the bokeh backend uses those values as frame_width/frame_height instead. This ensures the aspect is respected, but means that the plot might be slightly larger than anticipated. Set the frame_width/frame_height explicitly to suppress this warning.
WARNING:param.HeatMapPlot61028: Due to internal constraints, when aspect and width/height is set, the bokeh backend uses those values as frame_width/frame_height instead. This ensures the aspect is respected, but means that the plot might be slightly larger than anticipated. Set the frame_width/frame_height explicitly to suppress this warning.
WARNING:param.HeatMapPlot61130: Due to internal constraints, when aspect and width/height is set, the bokeh backend uses those values as frame_width/frame_height instead. This ensures the aspect is respected, but means that the plot might be slightly larger than anticipated. Set the frame_width/frame_height explicitly to suppress this warning.
WARNING:param.HeatMapPlot61165: Due to internal constraints, when aspect and width/height is set, the bokeh backend uses those values as frame_width/frame_height instead. This ensures the aspect is respected, but means that the plot might be slightly larger than anticipated. Set the frame_width/frame_height explicitly to suppress this warning.
WARNING:param.HeatMapPlot61266: Due to internal constraints, when aspect and width/height is set, the bokeh backend uses those values as frame_width/frame_height instead. This ensures the aspect is respected, but means that the plot might be slightly larger than anticipated. Set the frame_width/frame_height explicitly to suppress this warning.
WARNING:param.HeatMapPlot61301: Due to internal constraints, when aspect and width/height is set, the bokeh backend uses those values as frame_width/frame_height instead. This ensures the aspect is respected, but means that the plot might be slightly larger than anticipated. Set the frame_width/frame_height explicitly to suppress this warning.
Versions
[47]:
import scooby
scooby.Report(additional=['epsproc', 'pemtk','holoviews', 'hvplot', 'xarray','numpy','pandas', 'matplotlib', 'bokeh'])
[47]:
| Fri Jul 29 11:30:10 2022 Eastern Daylight Time | |||||
| OS | Windows | CPU(s) | 32 | Machine | AMD64 |
| Architecture | 64bit | RAM | 63.9 GB | Environment | Jupyter |
| Python 3.7.3 (default, Apr 24 2019, 15:29:51) [MSC v.1915 64 bit (AMD64)] | |||||
| epsproc | 1.3.2-dev | pemtk | 0.0.1 | holoviews | 1.14.8 |
| hvplot | 0.6.0 | xarray | 0.15.0 | numpy | 1.18.1 |
| pandas | 1.0.1 | matplotlib | 3.3.1 | bokeh | 1.4.0 |
| scipy | 1.3.0 | IPython | 7.12.0 | scooby | 0.5.6 |
| Intel(R) Math Kernel Library Version 2020.0.0 Product Build 20191125 for Intel(R) 64 architecture applications | |||||
[48]:
# Check current Git commit for local ePSproc version
from pathlib import Path
!git -C {Path(ep.__file__).parent} branch
!git -C {Path(ep.__file__).parent} log --format="%H" -n 1
* dev
master
numba-tests
b3a28d11374935812dd9b837d3acc661ea26fc1f
[49]:
# Check current remote commits
!git ls-remote --heads https://github.com/phockett/ePSproc
3b16244eb24918d0ad57af9b3e8fa76e36903b0e refs/heads/dev
54b929025381f1c2cbf371180fa870f247e7f627 refs/heads/master
69cd89ce5bc0ad6d465a4bd8df6fba15d3fd1aee refs/heads/numba-tests
ea30878c842f09d525fbf39fa269fa2302a13b57 refs/heads/revert-9-master
[50]:
# Check current Git commit for local PEMtk version
from pathlib import Path
!git -C {Path(pemtk.__file__).parent} branch
!git -C {Path(pemtk.__file__).parent} log --format="%H" -n 1
* master
5fd0ade08cb745ab0a8acf183644c30e48c0fd07
[51]:
# Check current remote commits
!git ls-remote --heads https://github.com/phockett/pemtk
5fd0ade08cb745ab0a8acf183644c30e48c0fd07 refs/heads/master
[ ]: